Solid-state NMR studies of amyloid fibril structure - PubMed (original) (raw)
Review
Solid-state NMR studies of amyloid fibril structure
Robert Tycko. Annu Rev Phys Chem. 2011.
Abstract
Current interest in amyloid fibrils stems from their involvement in neurodegenerative and other diseases and from their role as an alternative structural state for many peptides and proteins. Solid-state nuclear magnetic resonance (NMR) methods have the unique capability of providing detailed structural constraints for amyloid fibrils, sufficient for the development of full molecular models. In this article, recent progress in the application of solid-state NMR to fibrils associated with Alzheimer's disease, prion fibrils, and related systems is reviewed, along with relevant developments in solid-state NMR techniques and technology.
Figures
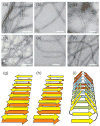
Figure 1
Negatively-stained TEM images of amyloid fibrils. (a) “Striated ribbon” Aβ1-40 fibrils. (b) “Twisted pair” Aβ1-40 fibrils. (c) Brain-seeded Aβ1-40 fibrils. (d) Amylin fibrils. (e) Sup35NM fibrils. (f) HET-s218-289 fibrils. White scale bars are 100 nm. (g,h,i) Schematic representations of cross-β structures formed by parallel β-sheets, antiparallel β-sheets, and a β-helix, respectively.
Figure 2
Molecular structural models for “striated ribbon” (a) and “twisted pair” (b) Aβ1-40 fibrils, developed primarily from solid state NMR data with additional constraints from electron microscopy. These models include residues 9-40, with hydrophobic, negatively charged, postively charged (including His), and polar (including Gly) residues colored green, red, blue, and magenta, respectively. Both models are viewed down the long axis of the fibril, with three repeats shown. The full Aβ1-40 sequence is shown below.
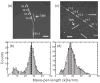
Figure 3
MPL data for Aβ1-40 fibrils. (a,c) Dark-field TEM images of unstained Aβ1-40 fibrils with “twisted pair” and “striated ribbon” morphologies, respectively. Arrows indicate tobacco mosaic virus (TMV) rods that serve as intensity calibration standards. Rectangles enclose fibril segments with the indicated MPL values in kDa/nm. White scale bars are 200 nm. (b,d) MPL histograms obtained from multiple dark-field images. Heavy dashed lines indicate the predicted MPL values for the 3-fold symmetric and 2-fold symmetric structural models in Figure 2, which are comprised of three and two cross-β units, respectively. Solid lines are multiple-Gaussian fits. Minor populations comprised of a single cross-β unit appear at MPL ≈ 9 kDa/nm, possibly arising from disruption of the fibril structures during TEM sample preparation or measurement.
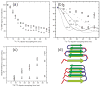
Figure 4
2D solid state NMR spectra of uniformly 15N,13C-labeled Aβ1-40 fibrils prepared by seeding with amyloid extracted from occipital lobe tissue of a deceased Alzheimer’s disease patient. Assignments of selected crosspeaks to specific residues are indicated. (a) 2D 13C-13C NMR spectrum, obtained in a 14.1 T magnetic field with 13.6 kHz MAS, using a 2.94 ms finite-pulse radio-frequency-driven recoupling (fpRFDR) sequence (117) for spin polarization transfer in the mixing period between the two spectral dimensions. (b) 2D 15N-13C spectrum, obtained with frequency-selective 15N-13C cross-polarization followed by fpRFDR in the mixing period.
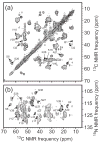
Figure 5
Experimental determination of β-sheet organization in wild-type and D23N mutant Aβ1-40 fibrils with 13C and 15N labels at specific sites. (a) Measurements of intermolecular 13C-13C dipole-dipole couplings in wild-type fibrils using the PITHIRDS-CT technique (98). Squares, circles, and triangles are data for A30 methyl distances in “striated ribbon” fibrils, A30 methyl distances in “twisted pair” fibrils, and M35 methyl distances in “twisted pair” fibrils. Decay of 13C NMR signals on the 30 ms time scale results from ~5 Å intermolecular 13C-13C distances, indicating an in-register parallel β-sheet structure. (b) Measurements of intermolecular couplings among A21 methyl carbons in D23N-Aβ1-40 fibrils containing parallel (triangles) and antiparallel (circles and squares) β-sheet structures. (c) Measurements of intermolecular 15N-13C dipole-dipole couplings between the backbone amide nitrogen of L17 and the methyl carbon of A21 in D23N-Aβ1-40 fibrils containing parallel (triangles) and antiparallel (circles and squares) β-sheet structures, using the REDOR technique (106). Build-up of the normalized REDOR difference signal ΔS/S0 on the 50 ms time scale indicates ~5 Å L17-A21 distances, consistent with an antiparallel β-sheet structure with 17+k ↔ 21-k hydrogen bond registry. (d) Schematic representation of cross-β units in which the two hydrophobic segments of Aβ1-40 (dark and light green arrows) form antiparallel (top) and parallel (bottom) β-sheets. Favorable hydrophobic interactions can occur in both structures.
Figure 6
Comparison of the HET-s218-289 prion fibril structure (a,b) with that of the β-helical protein Bordetella pertussis pertactin (c,d), based on Protein Data Bank files 2RNM and 1DAB, respectively. Structures are viewed perpendicular (a,c) and parallel (b,d) to the long axis of the fibril or β-helix. For HET-s218-289, five repeats of residues 226-278 are shown, with each repeat forming two “rungs” of the β-helix. For pertactin, residues 1-285 is shown. Hydrophobic, negatively charged, postively charged (including His), and polar (including Gly) residues are colored green, red, blue, and magenta, respectively.
Similar articles
- Molecular structures of amyloid and prion fibrils: consensus versus controversy.
Tycko R, Wickner RB. Tycko R, et al. Acc Chem Res. 2013 Jul 16;46(7):1487-96. doi: 10.1021/ar300282r. Epub 2013 Jan 7. Acc Chem Res. 2013. PMID: 23294335 Free PMC article. Review. - Supramolecular structural constraints on Alzheimer's beta-amyloid fibrils from electron microscopy and solid-state nuclear magnetic resonance.
Antzutkin ON, Leapman RD, Balbach JJ, Tycko R. Antzutkin ON, et al. Biochemistry. 2002 Dec 24;41(51):15436-50. doi: 10.1021/bi0204185. Biochemistry. 2002. PMID: 12484785 - Seeded growth of beta-amyloid fibrils from Alzheimer's brain-derived fibrils produces a distinct fibril structure.
Paravastu AK, Qahwash I, Leapman RD, Meredith SC, Tycko R. Paravastu AK, et al. Proc Natl Acad Sci U S A. 2009 May 5;106(18):7443-8. doi: 10.1073/pnas.0812033106. Epub 2009 Apr 17. Proc Natl Acad Sci U S A. 2009. PMID: 19376973 Free PMC article. - Structural variation in amyloid-β fibrils from Alzheimer's disease clinical subtypes.
Qiang W, Yau WM, Lu JX, Collinge J, Tycko R. Qiang W, et al. Nature. 2017 Jan 12;541(7636):217-221. doi: 10.1038/nature20814. Epub 2017 Jan 4. Nature. 2017. PMID: 28052060 Free PMC article. - Molecular structure of amyloid fibrils: insights from solid-state NMR.
Tycko R. Tycko R. Q Rev Biophys. 2006 Feb;39(1):1-55. doi: 10.1017/S0033583506004173. Epub 2006 Jun 13. Q Rev Biophys. 2006. PMID: 16772049 Review.
Cited by
- Accelerated Alzheimer's Aβ-42 secondary nucleation chronologically visualized on fibril surfaces.
Nirmalraj PN, Bhattacharya S, Thompson D. Nirmalraj PN, et al. Sci Adv. 2024 Oct 25;10(43):eadp5059. doi: 10.1126/sciadv.adp5059. Epub 2024 Oct 25. Sci Adv. 2024. PMID: 39454002 Free PMC article. - High resolution structural characterization of Aβ42 amyloid fibrils by magic angle spinning NMR.
Colvin MT, Silvers R, Frohm B, Su Y, Linse S, Griffin RG. Colvin MT, et al. J Am Chem Soc. 2015 Jun 17;137(23):7509-18. doi: 10.1021/jacs.5b03997. Epub 2015 Jun 4. J Am Chem Soc. 2015. PMID: 26001057 Free PMC article. - Yeast prions: structure, biology, and prion-handling systems.
Wickner RB, Shewmaker FP, Bateman DA, Edskes HK, Gorkovskiy A, Dayani Y, Bezsonov EE. Wickner RB, et al. Microbiol Mol Biol Rev. 2015 Mar;79(1):1-17. doi: 10.1128/MMBR.00041-14. Microbiol Mol Biol Rev. 2015. PMID: 25631286 Free PMC article. Review. - Utilizing afterglow magnetization from cross-polarization magic-angle-spinning solid-state NMR spectroscopy to obtain simultaneous heteronuclear multidimensional spectra.
Banigan JR, Traaseth NJ. Banigan JR, et al. J Phys Chem B. 2012 Jun 21;116(24):7138-44. doi: 10.1021/jp303269m. Epub 2012 May 31. J Phys Chem B. 2012. PMID: 22582831 Free PMC article. - Influence of Amino Acid Substitutions in ApoMb on Different Stages of Unfolding of Amyloids.
Katina N, Marchenkov V, Ryabova N, Ilyina N, Marchenko N, Balobanov V, Finkelstein A. Katina N, et al. Molecules. 2023 Nov 23;28(23):7736. doi: 10.3390/molecules28237736. Molecules. 2023. PMID: 38067466 Free PMC article.
References
- Sunde M, Blake CCF. From the globular to the fibrous state: Protein structure and structural conversion in amyloid formation. Q Rev Biophys. 1998;31:1–39. - PubMed
- Sacchettini JC, Kelly JW. Therapeutic strategies for human amyloid diseases. Nat Rev Drug Discov. 2002;1:267–75. - PubMed
- Chiti F, Dobson CM. Amyloid formation by globular proteins under native conditions. Nat Chem Biol. 2009;5:15–22. - PubMed
- Tycko R. Molecular structure of amyloid fibrils: Insights from solid state NMR. Q Rev Biophys. 2006;39:1–55. A comprehensive review of solid state NMR studies of amyloid fibrils and relevant methodology up to 2005. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources