SET8 is degraded via PCNA-coupled CRL4(CDT2) ubiquitylation in S phase and after UV irradiation - PubMed (original) (raw)
SET8 is degraded via PCNA-coupled CRL4(CDT2) ubiquitylation in S phase and after UV irradiation
Stine Jørgensen et al. J Cell Biol. 2011.
Abstract
The eukaryotic cell cycle is regulated by multiple ubiquitin-mediated events, such as the timely destruction of cyclins and replication licensing factors. The histone H4 methyltransferase SET8 (Pr-Set7) is required for chromosome compaction in mitosis and for maintenance of genome integrity. In this study, we show that SET8 is targeted for degradation during S phase by the CRL4(CDT2) ubiquitin ligase in a proliferating cell nuclear antigen (PCNA)-dependent manner. SET8 degradation requires a conserved degron responsible for its interaction with PCNA and recruitment to chromatin where ubiquitylation occurs. Efficient degradation of SET8 at the onset of S phase is required for the regulation of chromatin compaction status and cell cycle progression. Moreover, the turnover of SET8 is accelerated after ultraviolet irradiation dependent on the CRL4(CDT2) ubiquitin ligase and PCNA. Removal of SET8 supports the modulation of chromatin structure after DNA damage. These results demonstrate a novel regulatory mechanism, linking for the first time the ubiquitin-proteasome system with rapid degradation of a histone methyltransferase to control cell proliferation.
Figures
Figure 1.
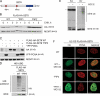
CRL-CDT2 targets SET8 for degradation in S phase. (A) SET8 protein level is low in S phase. U2-OS cells were treated with aphidicolin for 24 h and released for 4 h. The indicated cells were treated with MG132 1 h before harvest. Cells were processed for immunoblotting with the indicated antibodies. Vinculin was used as a loading control. Async, asynchronous control cells. Protein levels of SET8 are quantified and depicted below the figure. (B) SET8 contains a conserved PIP box degron. Cluster alignment of SET8 from different species show a highly conserved stretch of amino acids. When this stretch was aligned with CDT1 and p21, it revealed a conserved PIP box degron. (C) SET8 is mutually exclusive with PCNA and CDT2 on chromatin. U2-OS cells were synchronized with nocodazole, released into fresh medium, and collected at the indicated time points. Chromatin-bound fractions and whole cell extracts (WCE) were analyzed by immunoblotting with the indicated antibodies. (D) DDB1 and CDT2 depletion stabilizes SET8. U2-OS cells were treated with the indicated siRNAs 12 h before aphidicolin treatment. 24 h after aphidicolin treatment, cells were washed and released into fresh medium for 3 h and processed for immunoblotting with the indicated antibodies. Actin was used as a loading control. (E) CDT2 is important for the turnover of SET8 in S phase. U2-OS cells were treated with siRNA 12 h before aphidicolin treatment. 24 h after aphidicolin treatment, cells were washed and released into fresh medium for 3 h. Cells were then treated with cycloheximide (CHX) as indicated and processed for immunoblotting with the indicated antibodies. The blots were quantified using an image reader. Representative data from three similar experiments are shown. Data are presented as mean + SD. (F) SET8 is ubiquitylated in vivo mediated by CDT2. HEK293 cells were depleted for CDT2 using siRNA 12 h before transfection with Flag-HA-SET8 and HIS-ubiquitin (HIS-UBQ) as indicated. Cells were processed for immunoprecipitation using cobalt beads and immunoblotted with the indicated antibodies.
Figure 2.
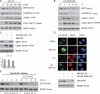
The interaction between PCNA and SET8 is important for CDT2-mediated degradation of SET8. (A) Schematic presentation of SET8. Two PIP boxes (PCNA-interacting domains) have been identified N terminally to the SET domain. (B) Mutation of PIP2 results in stabilization of SET8. HEK293 cells were transfected with plasmids expressing Flag-HA-SET8, Flag-HA-SET8-*PIP1, and Flag-HA-SET8-*PIP2. Cells were treated with cycloheximide and processed for immunoblotting with the indicated antibodies. MCM7 was used as a loading control. (C) Increased levels of PCNA result in degradation of WT SET8 but not SET8-*PIP2. HEK293 cells were transfected as illustrated and processed for immunoblotting with the indicated antibodies. (D) In vivo ubiquitylation of SET8-*PIP2 is reduced compared with SET8 WT. HEK293 cells were transfected with Flag-HA-SET8, Flag-HA-SET8-*PIP2, and HIS-ubiquitin as indicated. Cells were processed for immunoprecipitation using cobalt beads and immunoblotted with the indicated antibodies. (E) SET8 is ubiquitylated on chromatin. U2-OS cells were either treated with MG132 for 3 h or left untreated. Soluble and chromatin-bound fractions were analyzed by immunoblotting with SET8 antibody. *, nonspecific band. (F) SET8 WT and PCNA colocalize on chromatin after MG132 treatment. U2-OS cells expressing Flag-HA-SET8 or Flag-HA-SET8-*PIP2 were seeded on coverslips and treated with MG132 for 3 h before preextraction and fixation. Cells were stained with antibodies against HA and PCNA. Bars, 10 µm.
Figure 3.
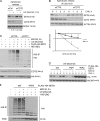
SET8 turnover is accelerated after UV damage, and SET8 is degraded after UV. (A) U2-OS cells were treated with increasing amounts of UV damage and processed for immunoblotting 3 h after treatment as indicated. (B) U2-OS cells were treated with a fixed dose of UV, harvested at the indicated time points, and processed for immunoblotting. (C and D) Proteasomal inhibition blocks SET8 degradation. (C) U2-OS cells were treated with UV alone or a combination of UV and MG132 for 1 h. Cells were processed for immunoblotting and quantitative PCR as indicated. Data are represented as mean + SD. (D) U2-OS cells were infected with an HA-SET8–expressing virus and seeded onto coverslips. Cells were treated with MG132, UV, or a combination 6 h before fixation and harvest. Coverslips were processed for immunofluorescence with the indicated antibodies. The remaining cells were processed for immunoblotting with the indicated antibodies. Bars, 10 µm. (E) UV treatment accelerates degradation of SET8. U2-OS cells were synchronized using aphidicolin and release into S phase, where they were treated with cycloheximide or a combination of cycloheximide and UV as indicated. Cells were processed for immunoblotting with the indicated antibodies.
Figure 4.
CRL4(CDT2) targets SET8 for degradation after UV. (A) CDT2 depletion stabilizes SET8 after UV. U2-OS cells were depleted for CDT2 and synchronized with aphidicolin. Cells were released into mid–S phase and treated with UV, harvested after 2 h, and processed for immunoblotting with the indicated antibodies. (B) Knockdown of CDT2 decreases SET8 turnover after UV. U2-OS cells were treated with siRNA 12 h before aphidicolin treatment. 24 h after aphidicolin treatment, cells were washed and released into fresh medium for 3 h. Cells were then treated with cycloheximide and UV as indicated and processed for immunoblotting with the indicated antibodies. The blots were quantified using an image reader. This is representative data from three similar experiments. Data are presented as mean + SD. (C) UV increases in vivo ubiquitylation of SET8. HEK293 cells were depleted for CDT2 using siRNA 8 h before transfection with Flag-HA-SET8 and HIS-ubiquitin as indicated. Cells were treated with UV or a combination of UV and MG132 2 h before harvest followed by immunoprecipitation using cobalt beads and immunoblotted with the indicated antibodies. (D) Mutation of PIP2 but not PIP1 increases stability of SET8 after UV. HEK293 cells were transfected with either Flag-HA-SET8, -*PIP1, or -*PIP2. They were treated with a combination of cycloheximide and UV or cycloheximide, UV, and MG132. Cells were processed for immunoblotting with the indicated antibodies. (E) WT SET8 but not SET8-*PIP2 is ubiquitylated in vivo after UV treatment. Cells were transfected with Flag-HA-SET8, Flag-HA-SET8-*PIP2, and HIS-ubiquitin as indicated. Cells were processed for immunoprecipitation using cobalt beads and immunoblotted with the indicated antibodies.
Figure 5.
Degradation of SET8 allows cell cycle progression beyond G1 phase. (A) Nondegradable SET8 reduces entry into mitosis. U2-OS cells were cotransfected with H2B-GFP and the indicated Flag-HA-SET8 constructs in a 1:3 ratio and simultaneously treated with aphidicolin for 24 h. Cells were released into fresh medium containing nocodazole, harvested, and fixed at the indicated time points. Cells were stained with H3S10P antibody, and cells positive for both GFP and H3S10P were identified by flow cytometry. Data are presented as mean + SD. (B) Expression levels of SET8 constructs. U2-OS cells were treated with nocodazole for 16 h (Noco) or transfected with the indicated pBabe or pCMV constructs 36 h before harvest. Cells were processed for immunoblotting with the indicated antibodies. Only 0.1 of the pCMV SET8 WT lysate was loaded compared with pBabe SET8 lysates. (C) SET8 WT and SET8-*PIP2 expression induces compaction of chromatin. pCMV Flag-HA-SET8 constructs were expressed in U2-OS cells seeded on coverslips and simultaneously treated with aphidicolin for 24 h. Cells were released into fresh medium and fixed after 4 h. Chromatin structure in SET8-expressing cells was analyzed by immunofluorescence stainings for DAPI in HA-positive cells. Bars, 10 µm. (D) Elevated levels of SET8-*PIP2 but not SET8-*PIP2/*SET results in a more compact chromatin structure. HEK293 cells were transfected with either Flag-HA-SET8-*PIP2 or Flag-HA-SET8-*PIP2/*SET constructs. Cells were harvested for MNase digest followed by separation of the DNA on an agarose gel. (E) Elevated levels of SET8-*PIP2 but not SET8-*PIP2/*SET results in a more compact chromatin structure in response to UV. HEK293 cells were transfected with either Flag-HA-SET8-*PIP2 or Flag-HA-SET8-*PIP2/*SET constructs. Cells were treated with UV (100 J/m2) 3 h before harvest. Cells were harvested for MNase digest followed by separation of the DNA on an agarose gel.
Similar articles
- CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase.
Centore RC, Havens CG, Manning AL, Li JM, Flynn RL, Tse A, Jin J, Dyson NJ, Walter JC, Zou L. Centore RC, et al. Mol Cell. 2010 Oct 8;40(1):22-33. doi: 10.1016/j.molcel.2010.09.015. Mol Cell. 2010. PMID: 20932472 Free PMC article. - Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage.
Oda H, Hübner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, Reinberg D. Oda H, et al. Mol Cell. 2010 Nov 12;40(3):364-76. doi: 10.1016/j.molcel.2010.10.011. Epub 2010 Oct 28. Mol Cell. 2010. PMID: 21035370 Free PMC article. - CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation.
Abbas T, Shibata E, Park J, Jha S, Karnani N, Dutta A. Abbas T, et al. Mol Cell. 2010 Oct 8;40(1):9-21. doi: 10.1016/j.molcel.2010.09.014. Mol Cell. 2010. PMID: 20932471 Free PMC article. - Mechanism of CRL4(Cdt2), a PCNA-dependent E3 ubiquitin ligase.
Havens CG, Walter JC. Havens CG, et al. Genes Dev. 2011 Aug 1;25(15):1568-82. doi: 10.1101/gad.2068611. Genes Dev. 2011. PMID: 21828267 Free PMC article. Review. - CRL4Cdt2 Ubiquitin Ligase, A Genome Caretaker Controlled by Cdt2 Binding to PCNA and DNA.
Mazian MA, Yamanishi K, Rahman MZA, Ganasen M, Nishitani H. Mazian MA, et al. Genes (Basel). 2022 Jan 29;13(2):266. doi: 10.3390/genes13020266. Genes (Basel). 2022. PMID: 35205311 Free PMC article. Review.
Cited by
- DCAF14 regulates CDT2 to promote SET8-dependent replication fork protection.
Tirado-Class N, Hathaway C, Nelligan A, Nguyen T, Dungrawala H. Tirado-Class N, et al. Life Sci Alliance. 2023 Nov 8;7(1):e202302230. doi: 10.26508/lsa.202302230. Print 2024 Jan. Life Sci Alliance. 2023. PMID: 37940188 Free PMC article. - Direct role for proliferating cell nuclear antigen in substrate recognition by the E3 ubiquitin ligase CRL4Cdt2.
Havens CG, Shobnam N, Guarino E, Centore RC, Zou L, Kearsey SE, Walter JC. Havens CG, et al. J Biol Chem. 2012 Mar 30;287(14):11410-21. doi: 10.1074/jbc.M111.337683. Epub 2012 Feb 2. J Biol Chem. 2012. PMID: 22303007 Free PMC article. - Regulation of Mammalian DNA Replication via the Ubiquitin-Proteasome System.
Abbas T, Dutta A. Abbas T, et al. Adv Exp Med Biol. 2017;1042:421-454. doi: 10.1007/978-981-10-6955-0_19. Adv Exp Med Biol. 2017. PMID: 29357069 Free PMC article. Review. - DDB2 regulates DNA replication through PCNA-independent degradation of CDT2.
Wu X, Yu M, Zhang Z, Leng F, Ma Y, Xie N, Lu F. Wu X, et al. Cell Biosci. 2021 Feb 8;11(1):34. doi: 10.1186/s13578-021-00540-5. Cell Biosci. 2021. PMID: 33557942 Free PMC article. - Methylation multiplicity and its clinical values in cancer.
Dai X, Ren T, Zhang Y, Nan N. Dai X, et al. Expert Rev Mol Med. 2021 Mar 31;23:e2. doi: 10.1017/erm.2021.4. Expert Rev Mol Med. 2021. PMID: 33787478 Free PMC article. Review.
References
- Centore R.C., Havens C.G., Manning A.L., Li J.M., Flynn R.L., Tse A., Jin J., Dyson N.J., Walter J.C., Zou L. 2010. CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol. Cell. 40:22–33 10.1016/j.molcel.2010.09.015 - DOI - PMC - PubMed
- Fang J., Feng Q., Ketel C.S., Wang H., Cao R., Xia L., Erdjument-Bromage H., Tempst P., Simon J.A., Zhang Y. 2002. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr. Biol. 12:1086–1099 10.1016/S0960-9822(02)00924-7 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous