Alcohol alters DNA methylation patterns and inhibits neural stem cell differentiation - PubMed (original) (raw)
Alcohol alters DNA methylation patterns and inhibits neural stem cell differentiation
Feng C Zhou et al. Alcohol Clin Exp Res. 2011 Apr.
Abstract
Background: Potential epigenetic mechanisms underlying fetal alcohol syndrome (FAS) include alcohol-induced alterations of methyl metabolism, resulting in aberrant patterns of DNA methylation and gene expression during development. Having previously demonstrated an essential role for epigenetics in neural stem cell (NSC) development and that inhibiting DNA methylation prevents NSC differentiation, here we investigated the effect of alcohol exposure on genome-wide DNA methylation patterns and NSC differentiation.
Methods: Neural stem cells in culture were treated with or without a 6-hour 88 mM ("binge-like") alcohol exposure and examined at 48 hours, for migration, growth, and genome-wide DNA methylation. The DNA methylation was examined using DNA-methylation immunoprecipitation followed by microarray analysis. Further validation was performed using Independent Sequenom analysis.
Results: Neural stem cell differentiated in 24 to 48 hours with migration, neuronal expression, and morphological transformation. Alcohol exposure retarded the migration, neuronal formation, and growth processes of NSC, similar to treatment with the methylation inhibitor 5-aza-cytidine. When NSC departed from the quiescent state, a genome-wide diversification of DNA methylation was observed-that is, many moderately methylated genes altered methylation levels and became hyper- and hypomethylated. Alcohol prevented many genes from such diversification, including genes related to neural development, neuronal receptors, and olfaction, while retarding differentiation. Validation of specific genes by Sequenom analysis demonstrated that alcohol exposure prevented methylation of specific genes associated with neural development [cut-like 2 (cutl2), insulin-like growth factor 1 (Igf1), epidermal growth factor-containing fibulin-like extracellular matrix protein 1 (Efemp1), and SRY-box-containing gene 7 (Sox 7)]; eye development, lens intrinsic membrane protein 2 (Lim 2); the epigenetic mark Smarca2 (SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2); and developmental disorder [DiGeorge syndrome critical region gene 2 (Dgcr2)]. Specific sites altered by DNA methylation also correlated with transcription factor binding sites known to be critical for regulating neural development.
Conclusion: The data indicate that alcohol prevents normal DNA methylation programming of key neural stem cell genes and retards NSC differentiation. Thus, the role of DNA methylation in FAS warrants further investigation.
Copyright © 2011 by the Research Society on Alcoholism.
Figures
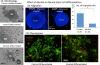
Figure 1. Alcohol treatment on Differentiation and Migration of neural stem cells
[1A] Brightfield Microscopy shows morphology of DRG neurospheres (NS) under three treatment groups: undifferentiated, differentiated, and alcohol-differentiated. Cells migrated out of neurosphere and grew processes as going through differentiation (Arrows). Alcohol reduced migration and fiber growth. DAPI staining (1B) shows less migration of differentiating cells in the alcohol-differentiated group (right, 1B and 1C) as compared with the control-differentiated group by student's t-test (left, IB and 1C) (*=p <0.05). 1D shows two phenotypes of the NSCs during differentiation. The MAP2-postive (MAP2+, green fluorescence, indicating neurons) cells some with fibers prevailed in the control differentiation (1D, left); the extend of MAP+ staining and fiber extension is apparently reduced when treated with alcohol (1D, right). Contrary to the MAP2+, the OCT4-positive (OCT4+, red fluorescence, indicating stem cells here) cells reduced through normal differentiation (1D, left), was held up when treated with alcohol (1D, right). Scale bars: 1A,100μm; 1B, 100μm; 1D, 50μm.
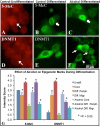
Figure 2. Immunostaining for 5-Methyl-cytidine (5-MeC) and DNMT1 epigenetic markers in neural stem cells during differentiation upon alcohol treatment
A-F shows translocation of epigenetic marks during differentiation. The 5-MeC immunoreactivity distributed throughout the nucleus (A, arrow) in undifferentiated NSCs, but relocated to the perinucleus (B, arrow) upon differentiation. Following alcohol treatment, 5-MeC was found not only in the perinucleus (C, arrow) but also regained nuclear-wide distribution (C, arrowhead). In undifferentiated NSCs, DNMT1 was expressed in the nucleus and cytoplasm (D, arrow); however, following differentiation, DNMT1 expression became exclusively cytoplasmic around nucleus (E). With alcohol treatment, DNMT1 was distributed in both the cytoplasm (F, arrowhead), and nucleus (F, arrows) which is normally seen in undifferentiated NSCs. Scale bars: A-F= 10um. A, D: Alexa633 (red fluorescence), and B, C, E, F: Alexa488 (Gree fluorescence). G, shows, through semiquantitative measurement, that 5-MeC and DNMT1 were dynamically changed from the undifferentiated control neurosphere (undiff.) to the differentiated Control and Alcohol groups in core and periphery (Periph.) of neurospheres and in migrated (Migr.) cells (n=10-12, *=p<0.05). The methods and statistics are described in the Methods.
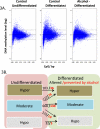
Figure 3. Genome-wide Distribution of NSC DNA methylation during differentiation and upon alcohol treatment
(3A) Scatter-plotting of promoter methylation levels against CpG content in NSC of three treatment groups demonstrated the pattern of DNA methylation. X-axis denotes the observed-to-expected CpG ratio, and Y-axis represents the average log2 transformation of the methylation signal level through all the probes in -1,300 to +500-bp from transcription start site. The dash lines at y=0.4 and -0.4 are defined as cutoff for hypermethylation (above upper line) and hypomethylation (below lower line), which represents DNA-methylation levels greater than 1.3 folds of the genome-wide median methylation level. The genes with DNA methylation levels between the two lines are defined as moderately methylated. (3B) shows the numbers of genes with DNA methylation changed during differentiation (Diff). The numbers in genes with DNA methylation changed during differentiation is indicated in black in three DNA methylation categories, and the number of genes prevented from change by alcohol during differentiation is shown in red and bold (p <0.05). For example, there were 103 hypermethylated genes that become more hypermethylated during differentiation, out of which 36 genes were prevented by alcohol; 222 moderate-methylated genes became hypermethylated, of which 78 were prevented (remained a moderate) by alcohol during differentiation.
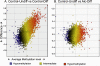
Figure 4. Genome-wide MvA plot compares DNA methylation difference between two groups
The difference of methylation between two groups is plotted on the Y-axis against the methylation level (X-axis, also indicated by color, see color legend). In (A) Control-Undifferentiated (Control-Undiff) vs. Control-Differentiated (Control-Diff), a large number of genes differ in methylation levels (Y-axis), and the degree of DNA methylation change is wide (X-axis). In contrast, in (B) Control-Undifferentiated (Control-Undiff) vs. Alcohol-Differentiated (Alc-Diff), the number of genes with methylation difference is reduced (Y-axis), and the degree of changes on DNA methylation level is smaller (X-axis).
Figure 5. Confirmation of alcohol-altered DNA methylation during differentiation by Sequenom analysis
Four genes, Igf1, Smarca2, Tnf, Cutl2 are shown as examples (in each gene, the Sequenom analysis is shown in upper, and MeDIP-Chip, lower). A blue dot indicates the CpG in control undifferentiated; a red diamond indicates CpG in Alcohol-treated. The red dashed lines indicate the significant region of change in methylation by MeDIP-chip.
Comment in
- Commentary: will analyzing the epigenome yield cohesive principles of ethanol teratology?
Miranda RC. Miranda RC. Alcohol Clin Exp Res. 2011 Jul;35(7):1201-3. doi: 10.1111/j.1530-0277.2011.01541.x. Epub 2011 May 9. Alcohol Clin Exp Res. 2011. PMID: 21554338 Review.
Similar articles
- Commentary: will analyzing the epigenome yield cohesive principles of ethanol teratology?
Miranda RC. Miranda RC. Alcohol Clin Exp Res. 2011 Jul;35(7):1201-3. doi: 10.1111/j.1530-0277.2011.01541.x. Epub 2011 May 9. Alcohol Clin Exp Res. 2011. PMID: 21554338 Review. - Chronic Ethanol Exposure Alters DNA Methylation in Neural Stem Cells: Role of Mouse Strain and Sex.
Amiri S, Davie JR, Rastegar M. Amiri S, et al. Mol Neurobiol. 2020 Feb;57(2):650-667. doi: 10.1007/s12035-019-01728-0. Epub 2019 Aug 14. Mol Neurobiol. 2020. PMID: 31414368 - Genome-Wide Transcriptome Landscape of Embryonic Brain-Derived Neural Stem Cells Exposed to Alcohol with Strain-Specific Cross-Examination in BL6 and CD1 Mice.
Xu W, Liyanage VRB, MacAulay A, Levy RD, Curtis K, Olson CO, Zachariah RM, Amiri S, Buist M, Hicks GG, Davie JR, Rastegar M. Xu W, et al. Sci Rep. 2019 Jan 18;9(1):206. doi: 10.1038/s41598-018-36059-y. Sci Rep. 2019. PMID: 30659253 Free PMC article. - Cellular DNA methylation program during neurulation and its alteration by alcohol exposure.
Zhou FC, Chen Y, Love A. Zhou FC, et al. Birth Defects Res A Clin Mol Teratol. 2011 Aug;91(8):703-15. doi: 10.1002/bdra.20820. Epub 2011 May 31. Birth Defects Res A Clin Mol Teratol. 2011. PMID: 21630420 - Gene methylation in gastric cancer.
Qu Y, Dang S, Hou P. Qu Y, et al. Clin Chim Acta. 2013 Sep 23;424:53-65. doi: 10.1016/j.cca.2013.05.002. Epub 2013 May 10. Clin Chim Acta. 2013. PMID: 23669186 Review.
Cited by
- Epigenetics and the power of art.
Karlic H, Baurek P. Karlic H, et al. Clin Epigenetics. 2011 Aug;2(2):279-82. doi: 10.1007/s13148-011-0033-7. Epub 2011 Apr 15. Clin Epigenetics. 2011. PMID: 22704342 Free PMC article. - Specific downregulation of microRNA-186 induces neural stem cell self-renewal by upregulating Bmi-1/FoxG1 expression.
Chen T, Liu J, Liu Y, Chen Y, Wang X. Chen T, et al. Hum Cell. 2023 Nov;36(6):2016-2026. doi: 10.1007/s13577-023-00981-9. Epub 2023 Sep 12. Hum Cell. 2023. PMID: 37700157 - The epigenetic landscape of alcoholism.
Krishnan HR, Sakharkar AJ, Teppen TL, Berkel TD, Pandey SC. Krishnan HR, et al. Int Rev Neurobiol. 2014;115:75-116. doi: 10.1016/B978-0-12-801311-3.00003-2. Int Rev Neurobiol. 2014. PMID: 25131543 Free PMC article. Review. - Alcohol exposure during development: Impact on the epigenome.
Perkins A, Lehmann C, Lawrence RC, Kelly SJ. Perkins A, et al. Int J Dev Neurosci. 2013 Oct;31(6):391-7. doi: 10.1016/j.ijdevneu.2013.03.010. Epub 2013 Mar 27. Int J Dev Neurosci. 2013. PMID: 23542005 Free PMC article.
References
- Abate P, Varlinskaya EI, Cheslock SJ, Spear NE, Molina JC. Neonatal activation of alcohol-related prenatal memories: impact on the first suckling response. Alcohol Clin Exp Res. 2002;26:1512–1522. - PubMed
- Bartolomei MS. Epigenetics: role of germ cell imprinting. Advances in Experimental Medicine and Biology. 2003;518:239–245. - PubMed
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. - PubMed
- Bonsch D, Lenz B, Fiszer R, Frieling H, Kornhuber J, Bleich S. Lowered DNA methyltransferase (DNMT-3b) mRNA expression is associated with genomic DNA hypermethylation in patients with chronic alcoholism. J Neural Transm. 2006;113:1299–1304. - PubMed
- Cheng LC, Tavazoie M, Doetsch F. Stem cells: from epigenetics to microRNAs. Neuron. 2005;46:363–367. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA085289/CA/NCI NIH HHS/United States
- R01 AA016698-03/AA/NIAAA NIH HHS/United States
- R01 AA016698-04/AA/NIAAA NIH HHS/United States
- P50 AA07611/AA/NIAAA NIH HHS/United States
- P60 AA007611-20/AA/NIAAA NIH HHS/United States
- P60 AA007611/AA/NIAAA NIH HHS/United States
- R01 AA016698/AA/NIAAA NIH HHS/United States
- P60 AA007611-19/AA/NIAAA NIH HHS/United States
- AA016698/AA/NIAAA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous