Multiplexed shotgun genotyping for rapid and efficient genetic mapping - PubMed (original) (raw)
Multiplexed shotgun genotyping for rapid and efficient genetic mapping
Peter Andolfatto et al. Genome Res. 2011 Apr.
Abstract
We present a new approach to genotyping based on multiplexed shotgun sequencing that can identify recombination breakpoints in a large number of individuals simultaneously at a resolution sufficient for most mapping purposes, such as quantitative trait locus (QTL) mapping and mapping of induced mutations. We first describe a simple library construction protocol that uses just 10 ng of genomic DNA per individual and makes the approach accessible to any laboratory with standard molecular biology equipment. Sequencing this library results in a large number of sequence reads widely distributed across the genomes of multiplexed bar-coded individuals. We develop a Hidden Markov Model to estimate ancestry at all genomic locations in all individuals using these data. We demonstrate the utility of the approach by mapping a dominant marker allele in D. simulans to within 105 kb of its true position using 96 F1-backcross individuals genotyped in a single lane on an Illumina Genome Analyzer. We further demonstrate the utility of our method by genetically mapping more than 400 previously unassembled D. simulans contigs to linkage groups and by evaluating the quality of targeted introgression lines. At this level of multiplexing and divergence between strains, our method allows estimation of recombination breakpoints to a median of 38-kb intervals. Our analysis suggests that higher levels of multiplexing and/or use of strains with lower levels of divergence are practicable.
Figures
Figure 1.
The experimental and bioinformatic pipeline for MSG. (1) Genomic DNA is fragmented with a restriction enzyme (RE) that leaves “sticky ends.” (2) Individual bar-coded adaptors are ligated to these restriction fragments. (3) Samples are pooled and, (4) the ligation products are size selected, PCR-amplified, and (5) sequenced on an Illumina Genome Analyzer. (6) Reads from the sequencing run are parsed based by barcode. (7) Each read is mapped to each of two parental genomes (indicated as red and blue, respectively). (8) Ancestry of chromosomal segments (blue: homozygous for parent 1; red: homozygous for parent 2; no color: heterozygous) is estimated using a Hidden Markov Model (HMM). (9) Genotypes and recombination breakpoints are used in downstream analyses, such as QTL mapping.
Figure 2.
Genome-wide ancestry assignment for a representative individual. (A) The ancestry states are shown for all major chromosome arms for a representative (male) individual progeny from a (D. sechellia/D. simulans) F1 X D. sechellia backcross experiment. The posterior probability that a region is homozygous for D. simulans (red) or for D. sechellia (blue) ancestry is plotted along the y axis. A high probability of heterozygous ancestry is indicated as a solid black line across the center of the plot. (B–D) Closer examination of three breakpoints illustrates typical variation in breakpoint resolution. Gold shading represents the 95% confidence bounds on the position of crossovers and the coordinates for each of these bounds are shown at the bottom.
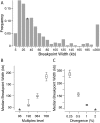
Figure 3.
Resolution of recombination breakpoints. (A) A histogram of 373 inferred recombination breakpoint intervals (coordinates where posterior probability of a given ancestry switches from ≥95% to ≤5%) for our backcross experiment. The red asterisks indicate the median breakpoint resolution in our experiment (96-plex, 2% divergence). (B,C) Box and whisker plots of the medians of 100 subsamples of the data to examine effects of (B) increased multiplexing and (C) decreased divergence between the strains being crossed.
Figure 4.
QTL map of the location of a dominant marker segregating in the reported backcross experiment. The inset illustrates below the estimated ancestries for all individuals between genomic locations 5.5 Mb and 8.5 Mb on the X chromosome and, above, the LOD profile. Individual ancestry estimates are sorted into individuals without EYFP above and with EYFP below. Regions with posterior probabilities close to 1 of homozygous D. simulans are coded blue, and homozygous D. sechellia are coded red. Posterior probabilities between 0 and 1 are coded with colors intermediate between blue and red.
Figure 5.
Three representative individuals from an experiment involving targeted introgression of D. simulans genomic regions into a D. sechellia genomic background. The color scheme is the same as in Figure 2. Individuals in A and B carry an introgression of a genomic region on 3R that was targeted with a dominant marker located at 9,390,800 bp on chromosome 3R (DL Stern, unpubl.), but also carry regions with D. simulans ancestry on the X and the tip of 2L, respectively (arrows). (C) Introgression of a region on 2L that was targeted with a dominant marker located at 5,926,416 bp on chromosome 2L (DL Stern, unpubl.) with no residual regions of D. simulans ancestry.
Similar articles
- A Major Locus Controls a Genital Shape Difference Involved in Reproductive Isolation Between Drosophila yakuba and Drosophila santomea.
Peluffo AE, Nuez I, Debat V, Savisaar R, Stern DL, Orgogozo V. Peluffo AE, et al. G3 (Bethesda). 2015 Oct 28;5(12):2893-901. doi: 10.1534/g3.115.023481. G3 (Bethesda). 2015. PMID: 26511499 Free PMC article. - Enhancing genetic mapping of complex genomes through the design of highly-multiplexed SNP arrays: application to the large and unsequenced genomes of white spruce and black spruce.
Pavy N, Pelgas B, Beauseigle S, Blais S, Gagnon F, Gosselin I, Lamothe M, Isabel N, Bousquet J. Pavy N, et al. BMC Genomics. 2008 Jan 18;9:21. doi: 10.1186/1471-2164-9-21. BMC Genomics. 2008. PMID: 18205909 Free PMC article. - Construction of a high-density genetic map by specific locus amplified fragment sequencing (SLAF-seq) and its application to Quantitative Trait Loci (QTL) analysis for boll weight in upland cotton (Gossypium hirsutum.).
Zhang Z, Shang H, Shi Y, Huang L, Li J, Ge Q, Gong J, Liu A, Chen T, Wang D, Wang Y, Palanga KK, Muhammad J, Li W, Lu Q, Deng X, Tan Y, Song W, Cai J, Li P, Rashid Ho, Gong W, Yuan Y. Zhang Z, et al. BMC Plant Biol. 2016 Apr 11;16:79. doi: 10.1186/s12870-016-0741-4. BMC Plant Biol. 2016. PMID: 27067834 Free PMC article. - QTL mapping using high-throughput sequencing.
Jamann TM, Balint-Kurti PJ, Holland JB. Jamann TM, et al. Methods Mol Biol. 2015;1284:257-85. doi: 10.1007/978-1-4939-2444-8_13. Methods Mol Biol. 2015. PMID: 25757777 Review. - The expanding scope of DNA sequencing.
Shendure J, Lieberman Aiden E. Shendure J, et al. Nat Biotechnol. 2012 Nov;30(11):1084-94. doi: 10.1038/nbt.2421. Epub 2012 Nov 8. Nat Biotechnol. 2012. PMID: 23138308 Free PMC article. Review.
Cited by
- Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species.
Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. Peterson BK, et al. PLoS One. 2012;7(5):e37135. doi: 10.1371/journal.pone.0037135. Epub 2012 May 31. PLoS One. 2012. PMID: 22675423 Free PMC article. - SNP-skimming: A fast approach to map loci generating quantitative variation in natural populations.
Wessinger CA, Kelly JK, Jiang P, Rausher MD, Hileman LC. Wessinger CA, et al. Mol Ecol Resour. 2018 Nov;18(6):1402-1414. doi: 10.1111/1755-0998.12930. Epub 2018 Aug 8. Mol Ecol Resour. 2018. PMID: 30033616 Free PMC article. - Reliable genotyping of recombinant genomes using a robust hidden Markov model.
Campos-Martin R, Schmickler S, Goel M, Schneeberger K, Tresch A. Campos-Martin R, et al. Plant Physiol. 2023 May 31;192(2):821-836. doi: 10.1093/plphys/kiad191. Plant Physiol. 2023. PMID: 36946207 Free PMC article. - Transgenic tools for targeted chromosome rearrangements allow construction of balancer chromosomes in non-melanogaster Drosophila species.
Stern DL. Stern DL. G3 (Bethesda). 2022 Apr 4;12(4):jkac030. doi: 10.1093/g3journal/jkac030. G3 (Bethesda). 2022. PMID: 35143616 Free PMC article. - Are assortative mating and genital divergence driven by reinforcement?
Hollander J, Montaño-Rendón M, Bianco G, Yang X, Westram AM, Duvaux L, Reid DG, Butlin RK. Hollander J, et al. Evol Lett. 2018 Oct 16;2(6):557-566. doi: 10.1002/evl3.85. eCollection 2018 Dec. Evol Lett. 2018. PMID: 30564439 Free PMC article.
References
- Ashburner M 1989. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, et al. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218 - PubMed
- Coop G, Wen X, Ober C, Pritchard JK, Przeworski M 2008. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science 319: 1395–1398 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50 GM071508/GM/NIGMS NIH HHS/United States
- R01 GM063622/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- GM063622-06A1/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases