MicroRNA characterize genetic diversity and drug resistance in pediatric acute lymphoblastic leukemia - PubMed (original) (raw)
MicroRNA characterize genetic diversity and drug resistance in pediatric acute lymphoblastic leukemia
Diana Schotte et al. Haematologica. 2011 May.
Erratum in
- Haematologica. 2011 Aug;96(8):1240
Abstract
Background: MicroRNA regulate the activity of protein-coding genes including those involved in hematopoietic cancers. The aim of the current study was to explore which microRNA are unique for seven different subtypes of pediatric acute lymphoblastic leukemia.
Design and methods: Expression levels of 397 microRNA (including novel microRNA) were measured by quantitative real-time polymerase chain reaction in 81 cases of pediatric leukemia and 17 normal hematopoietic control cases.
Results: All major subtypes of acute lymphoblastic leukemia, i.e. T-cell, MLL-rearranged, TEL-AML1-positive, E2A-PBX1-positive and hyperdiploid acute lymphoblastic leukemia, with the exception of BCR-ABL-positive and 'B-other' acute lymphoblastic leukemias (defined as precursor B-cell acute lymphoblastic leukemia not carrying the foregoing cytogenetic aberrations), were found to have unique microRNA-signatures that differed from each other and from those of healthy hematopoietic cells. Strikingly, the microRNA signature of TEL-AML1-positive and hyperdiploid cases partly overlapped, which may suggest a common underlying biology. Moreover, aberrant down-regulation of let-7b (~70-fold) in MLL-rearranged acute lymphoblastic leukemia was linked to up-regulation of oncoprotein c-Myc (P(FDR)<0.0001). Resistance to vincristine and daunorubicin was characterized by an approximately 20-fold up-regulation of miR-125b, miR-99a and miR-100 (P(FDR)≤0.002). No discriminative microRNA were found for prednisolone response and only one microRNA was linked to resistance to L-asparaginase. A combined expression profile based on 14 microRNA that were individually associated with prognosis, was highly predictive of clinical outcome in pediatric acute lymphoblastic leukemia (5-year disease-free survival of 89.4%±7% versus 60.8±12%, P=0.001).
Conclusions: Genetic subtypes and drug-resistant leukemic cells display characteristic microRNA signatures in pediatric acute lymphoblastic leukemia. Functional studies of discriminative and prognostically important microRNA may provide new insights into the biology of pediatric acute lymphoblastic leukemia.
Figures
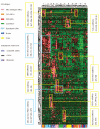
Figure 1.
Clustering of ALL subtypes and normal hematopoietic control cells by expression levels of 325 miRNA. Hierarchical clustering of ALL patients, normal bone marrow samples, CD34+ selected cells and thymocytes by expression levels of 325 (unselected) miRNA. Heatmap shows which miRNA are overex-pressed (in red) and which are underexpressed (in green) relative to snoRNA input control. Expression levels are plotted as standardized Z-scores per miRNA.
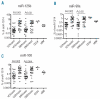
Figure 2.
Discriminative expression of miR-125b, miR-99a and miR-100 between drug-sensitive and -resistant precursor B-ALL patients. Expression levels relative to snoRNA are shown for miR-125b (A), miR-99a (B) and miR-100 (C). Dots represent individual samples of CD34+ selected cells (n=4), normal bone marrow samples ( =7) and the following precursor B-ALL patients: vincristine-sensitive (n=31; VCR/sens) and -resistant (n=30; VCR/resist) or daunorubicin-sensitive (n=29; DNR/sens) and -resistant (n=29; DNR/resist). Lines indicate median expression level in each group. The indicated P value is corrected for multiple-testing (FDR-corrected P value).
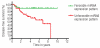
Figure 3.
Prognostic value of a miRNA expression profile in newly diagnosed pediatric ALL. Kaplan-Meier estimates for the probability of disease-free survival of a miRNA expression profile are shown. This profile comprises the combined score of 14 miRNA which were, independently of subtype, predictive for the clinical outcome of newly diagnosed pediatric ALL cases (see Table 3). See Design and Methods section for details on how this profile was generated. Basically, patients were assigned a score of 1 if the expression level of a given miRNA was associated with a good prognosis and patients were assigned a score 2 if this expression level was associated with a poor prognosis. Next, the sum of individual scores of 14 miRNA was taken which resulted in a minimum score of 14 and a maximum score of 28. Patients were divided in two groups based on the median score (of 21) in 78 patients and Kaplan-Meier estimates were calculated. _P_=0.001, Cox proportional hazard analysis.
Similar articles
- Expression of miR-196b is not exclusively MLL-driven but is especially linked to activation of HOXA genes in pediatric acute lymphoblastic leukemia.
Schotte D, Lange-Turenhout EA, Stumpel DJ, Stam RW, Buijs-Gladdines JG, Meijerink JP, Pieters R, Den Boer ML. Schotte D, et al. Haematologica. 2010 Oct;95(10):1675-82. doi: 10.3324/haematol.2010.023481. Epub 2010 May 21. Haematologica. 2010. PMID: 20494936 Free PMC article. - TEL/AML1 gene fusion is related to in vitro drug sensitivity for L-asparaginase in childhood acute lymphoblastic leukemia.
Ramakers-van Woerden NL, Pieters R, Loonen AH, Hubeek I, van Drunen E, Beverloo HB, Slater RM, Harbott J, Seyfarth J, van Wering ER, Hählen K, Schmiegelow K, Janka-Schaub GE, Veerman AJ. Ramakers-van Woerden NL, et al. Blood. 2000 Aug 1;96(3):1094-9. Blood. 2000. PMID: 10910927 - MiR-125b, miR-100 and miR-99a co-regulate vincristine resistance in childhood acute lymphoblastic leukemia.
Akbari Moqadam F, Lange-Turenhout EA, Ariës IM, Pieters R, den Boer ML. Akbari Moqadam F, et al. Leuk Res. 2013 Oct;37(10):1315-21. doi: 10.1016/j.leukres.2013.06.027. Epub 2013 Jul 31. Leuk Res. 2013. PMID: 23915977 - Pediatric acute lymphoblastic leukemia.
Carroll WL, Bhojwani D, Min DJ, Raetz E, Relling M, Davies S, Downing JR, Willman CL, Reed JC. Carroll WL, et al. Hematology Am Soc Hematol Educ Program. 2003:102-31. doi: 10.1182/asheducation-2003.1.102. Hematology Am Soc Hematol Educ Program. 2003. PMID: 14633779 Review. - Regulation of the miRNA expression by TEL/AML1, BCR/ABL, MLL/AF4 and TCF3/PBX1 oncoproteins in acute lymphoblastic leukemia (Review).
Organista-Nava J, Gómez-Gómez Y, Illades-Aguiar B, Leyva-Vázquez MA. Organista-Nava J, et al. Oncol Rep. 2016 Sep;36(3):1226-32. doi: 10.3892/or.2016.4948. Epub 2016 Jul 19. Oncol Rep. 2016. PMID: 27431573 Review.
Cited by
- Clinical significance of microRNAs in chronic and acute human leukemia.
Yeh CH, Moles R, Nicot C. Yeh CH, et al. Mol Cancer. 2016 May 14;15(1):37. doi: 10.1186/s12943-016-0518-2. Mol Cancer. 2016. PMID: 27179712 Free PMC article. Review. - Update on the Use of l-Asparaginase in Infants and Adolescent Patients with Acute Lymphoblastic Leukemia.
Andrade AF, Borges KS, Silveira VS. Andrade AF, et al. Clin Med Insights Oncol. 2014 Aug 24;8:95-100. doi: 10.4137/CMO.S10242. eCollection 2014. Clin Med Insights Oncol. 2014. PMID: 25210485 Free PMC article. Review. - The functional role of microRNA in acute lymphoblastic leukemia: relevance for diagnosis, differential diagnosis, prognosis, and therapy.
Luan C, Yang Z, Chen B. Luan C, et al. Onco Targets Ther. 2015 Oct 13;8:2903-14. doi: 10.2147/OTT.S92470. eCollection 2015. Onco Targets Ther. 2015. PMID: 26508875 Free PMC article. Review. - Integrative Analysis of MicroRNA-Mediated Gene Signatures and Pathways Modulating White Blood Cell Count in Childhood Acute Lymphoblastic Leukemia.
Ramani R, Megason G, Schallheim J, Karlson C, Vijayakumar V, Vijayakumar S, Hicks C. Ramani R, et al. Biomark Insights. 2017 Apr 12;12:1177271917702895. doi: 10.1177/1177271917702895. eCollection 2017. Biomark Insights. 2017. PMID: 28469402 Free PMC article.
References
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11(3):228–34. - PubMed
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303 (5654):83–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous