Crystal structures of complexes containing domains from two viral internal ribosome entry site (IRES) RNAs bound to the 70S ribosome - PubMed (original) (raw)
Crystal structures of complexes containing domains from two viral internal ribosome entry site (IRES) RNAs bound to the 70S ribosome
Jianyu Zhu et al. Proc Natl Acad Sci U S A. 2011.
Abstract
Internal ribosome entry site (IRES) RNAs are elements of viral or cellular mRNAs that bypass steps of canonical eukaryotic cap-dependent translation initiation. Understanding of the structural basis of IRES mechanisms is limited, partially due to a lack of high-resolution structures of IRES RNAs bound to their cellular targets. Prompted by the universal phylogenetic conservation of the ribosomal P site, we solved the crystal structures of proposed P site binding domains from two intergenic region IRES RNAs bound to bacterial 70S ribosomes. The structures show that these IRES domains nearly perfectly mimic a tRNA • mRNA interaction. However, there are clear differences in the global shape and position of this IRES domain in the intersubunit space compared to those of tRNA, supporting a mechanism for IRES action that invokes hybrid state mimicry to drive a noncanonical mode of translocation. These structures suggest how relatively small structured RNAs can manipulate complex biological machines.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
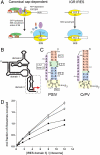
Translation initiation by the IGR IRESs and binding of domain 3 to the ribosome. (A) Comparison of canonical eukaryotic cap-dependent and IGR IRES-driven 80S ribosome recruitment. The IGR IRES does not use any protein factors or GTP hydrolysis. Protein initiation factors and the ribosomal subunits are depicted as colored ovals and labeled, and the initiator Met-tRNAmet is shown as a black vertical line. For clarity, an exhaustive list of factors involved in canonical initiation is not shown. (B) Secondary structure cartoon of the IGR IRES RNAs. The locations of the three domains that comprise the structure are shown, domain 3 is boxed, and the location of the start of protein synthesis is shown with a red arrow. (C) Secondary structures of the PSIV and CrPV IGR IRES domain 3 RNAs used in binding and crystallization studies. Elements of the structures are colored and labeled. The 3′-most nucleotides (lowercase) were mutated. (D) Binding of the PSIV (•,◯) and CrPV (▪,□) domain 3 RNAs shown in panel C to (•,▪) 70S or (◯,□) 80S ribosomes. See also
Figs. S1
and
S2
.
Fig. 2.
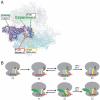
Structures of the PSIV and CrPV IGR IRES domain 3 RNAs bound to the T. thermophilus 70S ribosome. (A) σ_A-weighted 3Fo-2Fc electron density (contoured at 1.5σ of the PSIV domain 3 in the P site of the small subunit). (B) Placement of the PSIV domain 3 (red) within the ribosome structure. The ribosome is shown semitransparent to allow visualization of the IRES domain between the two subunits, which are labeled. (C) Comparison of the bound PSIV and CrPV domain 3 structures (Left and Right, colored and labeled to match Fig. 1_C) with the previously determined unbound CrPV domain 3 structure (Middle, cyan) (21). (D, Left and Middle) Interactions made between the PSIV and CrPV IRES domains (red) and the 16S rRNA (cyan). At right are the interactions between the 16S rRNA (cyan) and a P site-bound tRNA (orange, Right) paired to its cognate codon within a mRNA (green) (PDB ID code 2J00) (–29). Specific rRNA nucleotides listed in Table 1 are labeled. See also
Figs. S3
and
S4
.
Fig. 3.
Different trajectories of the IRES domain and tRNA. (A) Comparison of the interactions formed between a P site tRNA (Top, orange) and rRNA helix H69 (labeled), and the lack of interactions formed between the bound PSIV IRES domain 3 (red, Bottom) and H69. See also
Fig. S4
. (B, Top) Structure of a P/P state tRNA (orange), and mRNA (green) in the decoding groove of the 70S ribosome (gray) (28), showing a steric clash with a hybrid A/P state tRNA modeled into position (yellow). (Bottom) Our structure of the PSIV IRES domain 3 (red) in the P site, with a mRNA (green) and a modeled hybrid A/P state tRNA (yellow). The IRES domain would not sterically hinder movement of the A/A tRNA to an A/P hybrid state.
Fig. 4.
Proposed mechanism for the action of IGR IRES domain 3. (A) Results of superposition of the domain 3 RNA-70S ribosome structure presented here into the 7.3 Å cryo-EM density of a CrPV IGR IRES bound to an 80S ribosome (23), followed by docking of the PSIV IGR IRES domains 1 and 2 crystal structure (20) into the density (green). Only placement of domain 3 into the A site of the cryo-EM map matches the density (green) and is the proper distance from domains 1 and 2 to allow fitting of the 3-nucleotide linker (green). The location of an A site tRNA ASL (28) is shown (yellow). Placement of domain 3 into the P site (red) creates steric clash. See also
Fig. S5
. (B) Proposed mechanism for IGR IRES translocation (Bottom) compared to canonical translation initiation and translocation (Top). (Top) [A] The aminoacylated (orange) initiator P (cyan) and A site tRNAs (yellow) are bound to the ribosome (gray). [B] Peptidyl transfer results in a deacylated tRNA in the P site and a peptidyl tRNA in the A site. [C] The tRNAs sample the hybrid states. [D] The hybrid state is a substrate for eEF2-catalyzed translocation. (Bottom) [1] The IRES (green) assembles on an 80S ribosome. In the state shown, domain 3 is in the P site. [2] The orientation of domain 3 allows movement of the A site tRNA into an A/P hybrid state and this, combined with domains 1 and 2 of the IRES, promotes movement of the ribosome into a hybrid state. [3] An eEF2-catalyzed translocation event moves the IRES into the E site and the tRNA into the P site, completing the initiation process. Translation now can proceed through the normal elongation cycle.
Similar articles
- Initiation of translation in bacteria by a structured eukaryotic IRES RNA.
Colussi TM, Costantino DA, Zhu J, Donohue JP, Korostelev AA, Jaafar ZA, Plank TD, Noller HF, Kieft JS. Colussi TM, et al. Nature. 2015 Mar 5;519(7541):110-3. doi: 10.1038/nature14219. Epub 2015 Feb 4. Nature. 2015. PMID: 25652826 Free PMC article. - Structural basis for ribosome recruitment and manipulation by a viral IRES RNA.
Pfingsten JS, Costantino DA, Kieft JS. Pfingsten JS, et al. Science. 2006 Dec 1;314(5804):1450-4. doi: 10.1126/science.1133281. Epub 2006 Nov 23. Science. 2006. PMID: 17124290 Free PMC article. - tRNA-mRNA mimicry drives translation initiation from a viral IRES.
Costantino DA, Pfingsten JS, Rambo RP, Kieft JS. Costantino DA, et al. Nat Struct Mol Biol. 2008 Jan;15(1):57-64. doi: 10.1038/nsmb1351. Epub 2007 Dec 23. Nat Struct Mol Biol. 2008. PMID: 18157151 Free PMC article. - Comparing the three-dimensional structures of Dicistroviridae IGR IRES RNAs with other viral RNA structures.
Kieft JS. Kieft JS. Virus Res. 2009 Feb;139(2):148-56. doi: 10.1016/j.virusres.2008.07.007. Epub 2008 Aug 15. Virus Res. 2009. PMID: 18672012 Free PMC article. Review. - Viral IRES RNA structures and ribosome interactions.
Kieft JS. Kieft JS. Trends Biochem Sci. 2008 Jun;33(6):274-83. doi: 10.1016/j.tibs.2008.04.007. Epub 2008 May 28. Trends Biochem Sci. 2008. PMID: 18468443 Free PMC article. Review.
Cited by
- Processing and translation initiation of non-long terminal repeat retrotransposons by hepatitis delta virus (HDV)-like self-cleaving ribozymes.
Ruminski DJ, Webb CT, Riccitelli NJ, Lupták A. Ruminski DJ, et al. J Biol Chem. 2011 Dec 2;286(48):41286-41295. doi: 10.1074/jbc.M111.297283. Epub 2011 Oct 12. J Biol Chem. 2011. PMID: 21994949 Free PMC article. - Progress and outlook in structural biology of large viral RNAs.
Cantara WA, Olson ED, Forsyth KM. Cantara WA, et al. Virus Res. 2014 Nov 26;193:24-38. doi: 10.1016/j.virusres.2014.06.007. Epub 2014 Jun 21. Virus Res. 2014. PMID: 24956407 Free PMC article. Review. - Initiation of translation in bacteria by a structured eukaryotic IRES RNA.
Colussi TM, Costantino DA, Zhu J, Donohue JP, Korostelev AA, Jaafar ZA, Plank TD, Noller HF, Kieft JS. Colussi TM, et al. Nature. 2015 Mar 5;519(7541):110-3. doi: 10.1038/nature14219. Epub 2015 Feb 4. Nature. 2015. PMID: 25652826 Free PMC article. - Sordarin bound eEF2 unlocks spontaneous forward and reverse translocation on CrPV IRES.
Ou Z, Petrov A. Ou Z, et al. Nucleic Acids Res. 2023 Jul 21;51(13):6999-7013. doi: 10.1093/nar/gkad476. Nucleic Acids Res. 2023. PMID: 37283061 Free PMC article. - Taura syndrome virus IRES initiates translation by binding its tRNA-mRNA-like structural element in the ribosomal decoding center.
Koh CS, Brilot AF, Grigorieff N, Korostelev AA. Koh CS, et al. Proc Natl Acad Sci U S A. 2014 Jun 24;111(25):9139-44. doi: 10.1073/pnas.1406335111. Epub 2014 Jun 9. Proc Natl Acad Sci U S A. 2014. PMID: 24927574 Free PMC article.
References
- Pestova T, Lorsch JR, Hellen CU. In: Translational Control in Biology and Medicine. Mathews MB, Sonenberg N, Hershey JWB, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2007. pp. 87–128.
- Doudna JA, Sarnow P. In: Translational Control in Biology and Medicine. Mathews MB, Sonenberg N, Hershey JWB, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2007. pp. 129–153.
- Jan E. Divergent IRES elements in invertebrates. Virus Res. 2006;119:16–28. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM072560/GM/NIGMS NIH HHS/United States
- GM-17129/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- GM-59140/GM/NIGMS NIH HHS/United States
- R01 GM059140/GM/NIGMS NIH HHS/United States
- GM-72560/GM/NIGMS NIH HHS/United States
- GM-81346/GM/NIGMS NIH HHS/United States
- R37 GM017129/GM/NIGMS NIH HHS/United States
- R01 GM017129/GM/NIGMS NIH HHS/United States
- R01 GM081346/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous