Non-coding RNAs as regulators of embryogenesis - PubMed (original) (raw)
Review
Non-coding RNAs as regulators of embryogenesis
Andrea Pauli et al. Nat Rev Genet. 2011 Feb.
Abstract
Non-coding RNAs (ncRNAs) are emerging as key regulators of embryogenesis. They control embryonic gene expression by several means, ranging from microRNA-induced degradation of mRNAs to long ncRNA-mediated modification of chromatin. Many aspects of embryogenesis seem to be controlled by ncRNAs, including the maternal-zygotic transition, the maintenance of pluripotency, the patterning of the body axes, the specification and differentiation of cell types and the morphogenesis of organs. Drawing from several animal model systems, we describe two emerging themes for ncRNA function: promoting developmental transitions and maintaining developmental states. These examples also highlight the roles of ncRNAs in ensuring a robust commitment to one of two possible cell fates.
Figures
Figure 1. miR-430 — a multitasking microRNA family during embryogenesis
a | Clearance of maternal RNAs by miR-430. The miR-430 family promotes clearance of maternal RNAs in zebrafish and frogs. Maternal RNAs that are present in the egg drive early development in the absence of zygotic transcription. Activation of zygotic transcription leads to the expression of zygotic genes, including miR-430. Mature miR-430 accelerates the decay of hundreds of maternal RNAs. In the absence of Dicer (as in maternal-zygotic Dicer (MZ_dicer_) mutants), primary miR-430 transcripts are not processed into mature miR-430, which results in prolonged persistence of maternal RNAs. b | Modulation of miR-430 effector function. MicroRNAs (miRNAs) normally direct RNA-induced silencing complex (RISC) to the 3′ UTR of target genes and promote deadenylation of mRNAs (poly(A) tail shortening indicated by an arrow above AAAA). Binding of deleted in azoospermia-like (DAZL) to certain mRNAs antagonizes miRNA–RISC effector function by promoting polyadenylation of the mRNA. Similarly, binding of dead end 1 (DND1) to cis elements within 3′ UTRs of certain mRNAs prevents miRNA–RISC association. c | miR-430 family members regulate Nodal signalling. In fish and frogs, RISC, which contains miR-430/miR-427, dampens and balances Nodal signalling by repressing both agonistic (Nodal) and antagonistic (Lefty) ligands. The human orthologue of miR-430 (miR-302) targets only the antagonist LEFTY, and thereby enhances Nodal signalling. miRNA-mediated repression is shown in red (the thickness of the line indicates the strength of repression) and protein-mediated silencing or activation in black.
Figure 2. Regulation of pluripotency by microRNAs
Model for the various roles of different families of microRNAs (miRNAs) in the gene regulatory network that maintains pluripotent and differentiating cell states. Maintenance of embryonic stem (ES) cell fate depends on the activity of the ES cell-specific cell-cycle-regulating (ESCC) miRNAs, which are induced by the core pluripotency factors OCT4, Nanog, SOX2 and KLF4 or TCF3, as well as by MYC or MYCN. ESCC miRNAs trigger S phase entry by repressing negative G1–S phase regulators (for example, the serine–threonine protein kinase LATS2, the cyclin E-Cdk2 inhibitor p21 (also known as Dacapo) and retinoblastoma-like protein 2 (RBL2)). Key to the stabilization of either the pluripotent or the differentiated state is the antagonism between LIN28 (on in ES cells, off in differentiating cells) and let-7 (off in ES cells, on in differentiating cells). let-7 has a central role in promoting somatic differentiation by repressing multiple genes with important functions in ES cells. Other miRNAs that contribute to the suppression of pluripotent genes upon differentiation include miRNAs that repress pluripotency factors and miR-200 family members that repress the activity of Polycomb repressive complexes PRC1 and PRC2 (REFS 148,165,166). Red boxes indicate active miRNAs; unboxed text indicates inactive genes/miRNAs; grey lines indicate inactive processes. For further details, see the main text. Figure is modified, with permission, from REF. © (2010) Macmillan Publishing Ltd. All rights reserved.
Figure 3. RNAs control alternative cell fate decisions
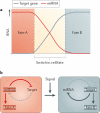
A recurrent theme of microRNA (miRNA) regulation during embryogenesis is their ability to control alternative cell fate decisions. Mutual repression between an miRNA and its target mRNA ensures reciprocal expression. The resulting bistable, double-negative feedback loop stabilizes either one of two possible cell fates and also promotes a rapid transition between the two states. Examples of bistable loops during embryogenesis include let-7–LIN28, miR-145–OCT4, miR-124–REST–SCP1, miR-9–TLX and miR-200–ZEB1.
Figure 4. Imprinting and dosage compensation
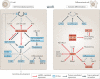
A | Imprinting. Parental-specific, monoallelic expression of gene clusters is based on differentially methylated imprinting control regions (ICRs). Only the unmethylated ICR (here shown on the paternal allele) is active and induces expression of a nearby long non-coding RNA (lncRNA, red). The lncRNA recruits repressive chromatin modifiers in cis to selected neighbouring genes, resulting in their silencing (OFF, dark blue). By contrast, a methylated ICR (here shown on the maternal allele) prevents expression of the lncRNA and thereby allows transcription of neighbouring genes (ON, light blue). B | Mechanism of X inactivation in mammals. In mammals, X chromosome inactivation occurs in distinct steps that depend on the activities of several lncRNAs that originate from the X-inactivation centre (Xic). Ba | Scheme of non-coding RNAs (ncRNAs) at the Xic locus. Bb | Steps of X inactivation in mammalian females. In embryonic stem (ES) cells, both X chromosomes express low levels of two key lncRNAs, Xist (red) and Tsix (green). Upon differentiation, one of the two X chromosomes is randomly selected to continue expressing Tsix (active X (Xa)). This process requires pairing, counting and choice (step 1). Tsix expression from Xa interferes with expression of Xist and RepA (yellow) in cis. On the future inactive X (Xi*), RepA recruits Polycomb repressive complex 2 (PRC2) to nucleate repressive chromatin marks, which are essential for upregulation of Xist expression on Xi* (step 2). Silencing spreads along the entire Xi* in an _Xist_-dependent manner, resulting in the establishment of a stable heterochromatic state on Xi (inactive X) (step 3). c | Comparison of dosage compensation in mammals and flies. In mammals, one of the two female X chromosomes is inactivated (dark blue), whereas flies upregulate the single X chromosome in males about twofold (green). ca | X inactivation in mammals depends on spreading of Xist (red) from its site of transcription along the entire length of the X chromosome. _Xist_-associated protein complexes such as PRC2, which catalyses trimethylation of lysine 27 on histone H3 (H3K27me3), establish a stably repressed chromatin state on Xi (dark blue). cb | By contrast, the fly dosage compensation complex (DCC), which contains the two ncRNAs roX1 and roX2 as structural components, binds discontinuously at hundreds of sites along the male X chromosome and deposits the activating H4K16-acetyl mark (twofold upregulated X chromosome in green). Panels Ba and Bb are modified, with permission, from REF. © (2010) Cold Spring Harbor Laboratory Press.
Figure 5. RNAs modulate chromatin
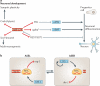
A | Models of gene regulation by _cis_- and _trans_-acting long non-coding RNAs (lncRNAs). Aa | In cis (left), the process of transcription can displace DNA-bound factors that inhibit (left) or activate (right) transcription of a neighbouring gene (process of transcription). Ab | Alternatively, nascent non-coding transcripts can function as tethers for chromatin-modifying complexes and/or transcriptional regulators, which can have either activating (left) or repressive (right) activities (tether model). Ac | _Trans_-acting non-coding RNAs (ncRNAs) can serve as platforms for the assembly of protein complexes (scaffold model). In this model, target sites are specified by DNA-binding proteins. Ad | Alternatively, _trans_-acting ncRNAs can specify target sites by forming hybrids with complementary DNA sequences, and thus recruit chromatin modifiers and transcriptional regulators (guide model). Ae | lncRNAs can also modulate the activity of protein complexes by inducing conformational changes (allosteric model). For simplicity, only repressive activities are shown as examples of _trans_-acting mechanisms. B | The lncRNA HOX antisense intergenic RNA (HOTAIR) regulates gene expression in trans by providing a scaffold for chromatin-modifying complexes. HOTAIR is expressed from the HOXC cluster and represses multiple target genes elsewhere in the genome. HOTAIR binds to the H3K27-trimethylating Polycomb repressive complex 2 (PRC2) and the H3K4me2/3-demethylating lysine-specific demethylase 1 (LSD1)–CoREST–REST complex, which together establish a repressive chromatin state at _HOTAIR_-associated target genes (OFF, dark blue). Pol II, RNA polymerase II.
Figure 6. Non-coding RNAs regulate neural development
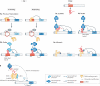
a | miR-124 promotes neural development by orchestrating the repression of several pathways that interfere with neural differentiation. Targeting of the mRNAs polypyrimidine tract-binding protein 1 (PTB), neuronal progenitor-specific BAF (npBAF) and small C-terminal domain phosphatase 1 (SCP1) by miR-124 promotes neural-specific splicing, neuronal chromatin remodelling and neural gene expression, respectively. These concerted changes are a driving force of neural differentiation. miR-124 has also been implicated in promoting adult neurogenesis in mice by repressing the transcription factor SOX9 (REF. 168). In Aplysia, miR-124 has been implicated in modulating synaptic plasticity by downregulating the transcriptional activator CREB. b | Cell fate specification by lsy-6 in Caenorhabditis elegans. A double-negative feedback loop under control of microRNAs and two transcription factors (DIE-1 and COG-1) specifies neuronal identities in C. elegans. Repression of cog-1 by DIE-1-induced lsy-6 is essential for establishing left side (ASEL) neuronal identities. Conversely, COG-1 interferes with DIE-1 expression in ASER neurons by activating a still unknown factor that represses die-1. Red boxes indicate active miRNAs; light blue boxes indicate active proteins; unboxed text indicates inactive elements; grey lines indicate inactive processes.
Figure 7. Regulation of the epithelial-to-mesenchymal transition by non-coding RNAs
Scheme of non-coding RNA (ncRNA)-mediated control of the epithelial-to-mesenchymal transition (EMT). miR-200 has a central role in stabilizing the epithelial state (green) by repressing negative regulators of E-cadherin expression. In epithelia, miR-200 targets subunits of Polycomb repressive complex 1 (PRC1) (BMI1) and PRC2 (SUZ12) as well as the transcription factors ZEB1 and ZEB2. Conversely, transforming growth factor-β (TGFβ)-induced ZEB1, ZEB2 and homologue of Snail (SNAI1) repress miR-200 and E-cadherin and promote mesenchymal fate (grey). Additional mechanisms that are implicated in inducing EMT include the positive-feedforward loop featuring the SNAI1-induced natural antisense transcript of ZEB2 (ZEB2-NAT) as well as TGFβ-induced maturation of primary miR-21 (pri-miR-21). For further details, see the main text. Red boxes indicate active miRNAs; light blue boxes indicate active proteins; unboxed text indicates inactive genes/miRNAs; grey lines indicate inactive processes. PDCD4, programmed cell death 4; PTEN, phosphatase and tensin homologue; MET, mesenchymal-to-epithelial transition; RISC, RNA-induced silencing complex.
Similar articles
- RNA-based regulation of pluripotency.
Wright JE, Ciosk R. Wright JE, et al. Trends Genet. 2013 Feb;29(2):99-107. doi: 10.1016/j.tig.2012.10.007. Epub 2012 Nov 10. Trends Genet. 2013. PMID: 23146412 Review. - Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation.
Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Soldà G, Simons C, Sunkin SM, Crowe ML, Grimmond SM, Perkins AC, Mattick JS. Dinger ME, et al. Genome Res. 2008 Sep;18(9):1433-45. doi: 10.1101/gr.078378.108. Epub 2008 Jun 18. Genome Res. 2008. PMID: 18562676 Free PMC article. - Single-Cell Non-coding RNA in Embryonic Development.
Fu Q, Liu CJ, Zhai ZS, Zhang X, Qin T, Zhang HW. Fu Q, et al. Adv Exp Med Biol. 2018;1068:19-32. doi: 10.1007/978-981-13-0502-3_3. Adv Exp Med Biol. 2018. PMID: 29943293 Review. - Non-coding RNAs: regulators of disease.
Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Taft RJ, et al. J Pathol. 2010 Jan;220(2):126-39. doi: 10.1002/path.2638. J Pathol. 2010. PMID: 19882673 Review. - Beyond mRNA: The role of non-coding RNAs in normal and aberrant hematopoiesis.
Wilkes MC, Repellin CE, Sakamoto KM. Wilkes MC, et al. Mol Genet Metab. 2017 Nov;122(3):28-38. doi: 10.1016/j.ymgme.2017.07.008. Epub 2017 Jul 25. Mol Genet Metab. 2017. PMID: 28757239 Free PMC article. Review.
Cited by
- MicroRNA-7 regulates the mTOR pathway and proliferation in adult pancreatic β-cells.
Wang Y, Liu J, Liu C, Naji A, Stoffers DA. Wang Y, et al. Diabetes. 2013 Mar;62(3):887-95. doi: 10.2337/db12-0451. Epub 2012 Dec 6. Diabetes. 2013. PMID: 23223022 Free PMC article. - PRKAR1B-AS2 Long Noncoding RNA Promotes Tumorigenesis, Survival, and Chemoresistance via the PI3K/AKT/mTOR Pathway.
Elsayed AM, Bayraktar E, Amero P, Salama SA, Abdelaziz AH, Ismail RS, Zhang X, Ivan C, Sood AK, Lopez-Berestein G, Rodriguez-Aguayo C. Elsayed AM, et al. Int J Mol Sci. 2021 Feb 13;22(4):1882. doi: 10.3390/ijms22041882. Int J Mol Sci. 2021. PMID: 33668685 Free PMC article. - The Potential Links between lncRNAs and Drug Tolerance in Lung Adenocarcinoma.
Davis WJH, Drummond CJ, Diermeier S, Reid G. Davis WJH, et al. Genes (Basel). 2024 Jul 11;15(7):906. doi: 10.3390/genes15070906. Genes (Basel). 2024. PMID: 39062685 Free PMC article. Review. - MicroRNAs in liver disease.
Wang XW, Heegaard NH, Orum H. Wang XW, et al. Gastroenterology. 2012 Jun;142(7):1431-43. doi: 10.1053/j.gastro.2012.04.007. Epub 2012 Apr 11. Gastroenterology. 2012. PMID: 22504185 Free PMC article. Review. - OGS2: genome re-annotation of the jewel wasp Nasonia vitripennis.
Rago A, Gilbert DG, Choi JH, Sackton TB, Wang X, Kelkar YD, Werren JH, Colbourne JK. Rago A, et al. BMC Genomics. 2016 Aug 25;17(1):678. doi: 10.1186/s12864-016-2886-9. BMC Genomics. 2016. PMID: 27561358 Free PMC article.
References
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. - PubMed
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. - PubMed
- Brown CJ, et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. - PubMed
- Brockdorff N, et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. - PubMed
- Borsani G, et al. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351:325–329. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources