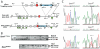Mouse survival motor neuron alleles that mimic SMN2 splicing and are inducible rescue embryonic lethality early in development but not late - PubMed (original) (raw)
Mouse survival motor neuron alleles that mimic SMN2 splicing and are inducible rescue embryonic lethality early in development but not late
Suzan M Hammond et al. PLoS One. 2010.
Abstract
Spinal muscular atrophy (SMA) is caused by low survival motor neuron (SMN) levels and patients represent a clinical spectrum due primarily to varying copies of the survival motor neuron-2 (SMN2) gene. Patient and animals studies show that disease severity is abrogated as SMN levels increase. Since therapies currently being pursued target the induction of SMN, it will be important to understand the dosage, timing and cellular requirements of SMN for disease etiology and potential therapeutic intervention. This requires new mouse models that can induce SMN temporally and/or spatially. Here we describe the generation of two hypomorphic Smn alleles, Smn(C-T-Neo) and Smn(2B-Neo). These alleles mimic SMN2 exon 7 splicing, titre Smn levels and are inducible. They were specifically designed so that up to three independent lines of mice could be generated, herein we describe two. In a homozygous state each allele results in embryonic lethality. Analysis of these mutants indicates that greater than 5% of Smn protein is required for normal development. The severe hypomorphic nature of these alleles is caused by inclusion of a loxP-flanked neomycin gene selection cassette in Smn intron 7, which can be removed with Cre recombinase. In vitro and in vivo experiments demonstrate these as inducible Smn alleles. When combined with an inducible Cre mouse, embryonic lethality caused by low Smn levels can be rescued early in gestation but not late. This provides direct genetic evidence that a therapeutic window for SMN inductive therapies may exist. Importantly, these lines fill a void for inducible Smn alleles. They also provide a base from which to generate a large repertoire of SMA models of varying disease severities when combined with other Smn alleles or SMN2-containing mice.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Generation of mutant Smn alleles.
(A) Gene targeting strategy to introduce the C-T and 2B mutation into Smn exon 7 using the gene targeting vectors pSmnC-T-Neo and pSmn2B-Neo. (B) Southern blot analysis of BamH I and Pst I digested DNA from neo resistant ES cell clones identified homologous recombinants. Two clones from each were used to perform blastocyst injections. (C) Germline transmission of SmnC-T-Neo and Smn2B-Neo alleles were determined by direct sequencing of Smn exon 7 PCR products from heterozygous mice. The C-T mutation corresponds to the nucleotide transition within exon 7 of the SMN2 gene. The 2B mutation corresponds to a mutation within the splice enhancer region 2B, changing GGA to TTT .
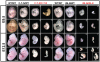
Figure 2. Whole mount analysis of embryos.
Smn2B-Neo or SmnC-T-Neo heterozygotes were intercrossed and embryos obtained at either E9.5 or E12.5 for whole-mount analysis and genotyping. (a–h) E9.5 SmnC-T-Neo embryos. Heterozygotes (C-T-N/WT) are identical to wild type (WT/WT) littermates. Homozygotes (C-T-N/C-T-N) are small but alive and larger than the Smn2B-Neo/2B-Neo (2B-N/2B-N) homozygotes. (i–p) E12.5 SmnC-T-Neo embryos. Homozygotes are extremely small compared to controls and many are being reabsorbed as shown in panel (p). (a'–h') E9.5 Smn2B-Neo embryos. Heterozygotes (2B-N/WT) are identical to wild type (WT/WT) littermates. Homozygotes (2B-N/2B-N) are developmentally retarded though still alive with signs of lethality clearly present before this period in some embryos that did not allow for genotyping (Table 1). (i'–p') E12.5 Smn2B-Neo embryos. All homozygous mutant embryos are undergoing resorption. Insets in o' and p' are magnified images of embryos in panel. Scale for all E9.5 embryos is 100 µM and for E12.5 200 µM.
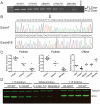
Figure 3. Smn transcript analysis analysis of E9.5 embryos.
(A) RT-PCR of E9.5 embryos comparing wild type, heterozygotes, and homozygotes. Both FL-Smn and Δ7Smn transcripts are amplified from cDNA of mice that are heterozygous and homozygous for the mutant alleles. (B) Direct sequencing of FL-Smn and Δ7Smn RT-PCR products derived from SmnC-T-Neo/C-T-Neo mutants. The “T” denoted with an arrow above it, represents the C-T mutation. Dotted line within exon6-8 sequence represents the junction between exon 6 and exon 8. (C) qRT-PCR results of E9.5 embryo for FL-Smn and Δ7Smn transcripts. FL-Smn transcripts for C-T-Neo and 2B-Neo were compared to wild type and heterozygous embryos within their own intercrosses and litters to control for variability. Spinal cord (S.C.) cDNA from a 6-month Δ7/WT mouse was used to compare Δ7Smn transcripts from C-T-Neo and 2B-Neo heterozygous and homozygous embryos. Wild type mice do not express Δ7Smn and are not shown on the graph. Each data point on the graphs represent individual embryos and depiction of variability between embryos. (D) Immunoblot analysis of Smn expression from individual E9.5 embryos derived from SmnC-T-Neo or Smn2B-Neo intercrosses. 15 times less protein was used for the controls to avoid overloading SMN while simultaneously detecting it in the mutants. Note the variation in Smn levels from individual mutant embryos. Lower Smn levels correlated with more severe phenotypes (see Figure 2). To achieve this sensitivity, Smn detection was performed on a Li-COR Odyssey infrared imaging system. Abbreviations: (WT) SmnWT/WT, (C-T-N/WT) SmnC-T-Neo/WT, (2B-N/WT) Smn2B-Neo/WT, (C-T-N/C-T-N) SmnC-T-Neo/C-T-Neo, (2B-N/2B-N) Smn2B-Neo/2B-Neo, (Δ7/WT) SmnΔ7/WT.
Figure 4. Smn expression is efficiently induced from the SmnC-T-Neo allele in vitro.
Two independent primary MEF cells lines were derived from double transgenic embryos (SmnC-T-Neo/WT;CreEsr1). (A) Schematic of the SmnC-T-Neo allele from exon 6 to exon 8. Arrows represent forward and reverse primers used in the 3-plex PCR reaction to identify SmnWT (640 & 637), SmnC-T-Neo (638 & 637), and SmnC-T (640 & 637) alleles. Primers 640 and 637 do not amplify a product in the presence of pgk-neo as the amplicon exceeds the time of elongation. (B) 3-plex PCR amplification of DNA from MEF lines 1 and 2 treated for 1 hr with 1 mM tamoxifen (+TM). MEF lines 1 and 2 left untreated (-TM) showed a slight amount of background excision (lanes 2 & 3); however, in the presence of tamoxifen (+TM), they readily amplify the SmnC-T allele (lanes 4&5). Controls in lanes 6, 7, and 8 were E10.5 embryos harvested to show the indicated genotypes from crosses using germline transmitting SmnC-T-Neo and SmnC-T alleles. (C) RNA from untreated (-TM) and induced (+TM) MEF cells were amplified by RT-PCR and FL-Smn transcripts directly sequenced. Induced MEFs (+TM) produced enough FL-Smn transcripts from the mutant C-T allele that could be detected by direct sequencing. The arrow points out the C-T mutation in +TM treated cultures. (D) qRT-PCR of FL-Smn and Δ7Smn from uninduced (-TM) and induced (+TM) cultures. Abbreviations: (TM) tamoxifen (C-T-N/WT;Cre+) SmnC-T-Neo/+;CreEsr1 (C-T-N/WT) SmnC-T-Neo/+ (C-T/WT) SmnC-T/+ (C-T-N/C-T) SmnC-T-Neo/C-T.
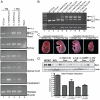
Figure 5. Smn induction in adults and embryos from a single injection of tamoxifen.
(A) DNA analysis of adult mice i.p. injected with vehicle (corn oil) or TM (9 mg/40 g body weight) using the same 3-plex PCR reaction as shown in Figures 4A and B. Wild type mice (WT;Cre-) only amplified the wild type allele (lane 1). Doubly transgenic mice (SmnC-T-Neo/WT;CreEsr1) in the absence of TM (-TM) displayed a low basal level of pgk-neo excision as has been previously reported for this Cre line . In the absence of the CreEsr1 transgene, SmnC-T-Neo/WT mice injected with TM could not excise pgk-neo (lane 3), in all tissues analyzed, pgk-neo excision was only possible and efficient in the presence of CreEsr1 and TM (lane 4). (B) PCR analysis of E18.5 embryos that received a single i.p. dose of TM (3 mg/40 g body weight) to the pregnant dam at E7.5 or E13.5 DNA was genotyped as above to differentiate SmnWT, SmnC-T-Neo and SmnC-T alleles. Arrows identify the appropriate amplicons. (C) Photomicrograph of E18.5 embryos induced with TM at E7.5 or E13.5. Lines in photograph show where images were tiled together in Photoshop. (D) Western blot and semi-quantitative densitometry of protein extracted from brain tissue of induced and control E18.5 embryos. A small amount of protein was able to be extracted from severely deformed SmnC-T-Neo/C-T-Neo embryos identified as “escapers” for comparison to induced SmnC-T-Neo/C-T-Neo;CreEsr1 rescued embryos. Semi-quantitative densitometry was performed on a separate blot using the same samples shown and normalized to β-tubulin, without the uninduced mutant. Protein levels from induced homozygous embryos, SmnC-T-Neo/C-T-Neo;CreESR1, (0.7±0.10) was greater than SmnWT/- (0.5±0.2). Abbreviations: (WT) Smn wild type allele, (Cre+ and Cre-) presence or absence of CreEsr1, (C-T-N/WT) SmnC-T-Neo/WT, (C-T-N/C-T-N) SmnC-T-Neo/C-T-Neo, (C-T) SmnC-T allele, (C-T-Neo) SmnC-T-Neo allele, (TM) tamoxifen.
Similar articles
- Deficiency of the splicing factor Sfrs10 results in early embryonic lethality in mice and has no impact on full-length SMN/Smn splicing.
Mende Y, Jakubik M, Riessland M, Schoenen F, Rossbach K, Kleinridders A, Köhler C, Buch T, Wirth B. Mende Y, et al. Hum Mol Genet. 2010 Jun 1;19(11):2154-67. doi: 10.1093/hmg/ddq094. Epub 2010 Feb 27. Hum Mol Genet. 2010. PMID: 20190275 - A humanized Smn gene containing the SMN2 nucleotide alteration in exon 7 mimics SMN2 splicing and the SMA disease phenotype.
Gladman JT, Bebee TW, Edwards C, Wang X, Sahenk Z, Rich MM, Chandler DS. Gladman JT, et al. Hum Mol Genet. 2010 Nov 1;19(21):4239-52. doi: 10.1093/hmg/ddq343. Epub 2010 Aug 12. Hum Mol Genet. 2010. PMID: 20705738 Free PMC article. - ZPR1 prevents R-loop accumulation, upregulates SMN2 expression and rescues spinal muscular atrophy.
Kannan A, Jiang X, He L, Ahmad S, Gangwani L. Kannan A, et al. Brain. 2020 Jan 1;143(1):69-93. doi: 10.1093/brain/awz373. Brain. 2020. PMID: 31828288 Free PMC article. - Is RNA manipulation a viable therapy for spinal muscular atrophy?
Horne C, Young PJ. Horne C, et al. J Neurol Sci. 2009 Dec 15;287(1-2):27-31. doi: 10.1016/j.jns.2009.08.055. Epub 2009 Sep 15. J Neurol Sci. 2009. PMID: 19758605 Review. - New and Developing Therapies in Spinal Muscular Atrophy: From Genotype to Phenotype to Treatment and Where Do We Stand?
Chen TH. Chen TH. Int J Mol Sci. 2020 May 7;21(9):3297. doi: 10.3390/ijms21093297. Int J Mol Sci. 2020. PMID: 32392694 Free PMC article. Review.
Cited by
- Mechanisms of exercise-induced survival motor neuron expression in the skeletal muscle of spinal muscular atrophy-like mice.
Ng SY, Mikhail A, Ljubicic V. Ng SY, et al. J Physiol. 2019 Sep;597(18):4757-4778. doi: 10.1113/JP278454. Epub 2019 Aug 22. J Physiol. 2019. PMID: 31361024 Free PMC article. - A short antisense oligonucleotide ameliorates symptoms of severe mouse models of spinal muscular atrophy.
Keil JM, Seo J, Howell MD, Hsu WH, Singh RN, DiDonato CJ. Keil JM, et al. Mol Ther Nucleic Acids. 2014 Jul 8;3(7):e174. doi: 10.1038/mtna.2014.23. Mol Ther Nucleic Acids. 2014. PMID: 25004100 Free PMC article. - Behavioral and electrophysiological outcomes of tissue-specific Smn knockdown in Drosophila melanogaster.
Timmerman C, Sanyal S. Timmerman C, et al. Brain Res. 2012 Dec 13;1489:66-80. doi: 10.1016/j.brainres.2012.10.035. Epub 2012 Oct 26. Brain Res. 2012. PMID: 23103409 Free PMC article. - DNA Damage Response and DNA Repair in Skeletal Myocytes From a Mouse Model of Spinal Muscular Atrophy.
Fayzullina S, Martin LJ. Fayzullina S, et al. J Neuropathol Exp Neurol. 2016 Sep;75(9):889-902. doi: 10.1093/jnen/nlw064. Epub 2016 Jul 24. J Neuropathol Exp Neurol. 2016. PMID: 27452406 Free PMC article. - The contribution of mouse models to understanding the pathogenesis of spinal muscular atrophy.
Sleigh JN, Gillingwater TH, Talbot K. Sleigh JN, et al. Dis Model Mech. 2011 Jul;4(4):457-67. doi: 10.1242/dmm.007245. Dis Model Mech. 2011. PMID: 21708901 Free PMC article. Review.
References
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, et al. Identification and characterization of a spinal muscular atrophy- determining gene. Cell. 1995;80:155–165. - PubMed
- Munsat TL, Davies KE. International SMA consortium meeting. (26–28 June 1992, Bonn, Germany). Neuromuscul Disord. 1992;2:423–428. - PubMed
- Wang CH, Finkel RS, Bertini ES, Schroth M, Simonds A, et al. Consensus statement for standard of care in spinal muscular atrophy. J Child Neurol. 2007;22:1027–1049. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R21 HD058311/HD/NICHD NIH HHS/United States
- CAPMC/ CIHR/Canada
- 1R01NS060926/NS/NINDS NIH HHS/United States
- T32 AG000260/AG/NIA NIH HHS/United States
- R01 NS060926/NS/NINDS NIH HHS/United States
- 1R21HD058311/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases