Factor-inhibiting hypoxia-inducible factor (FIH) catalyses the post-translational hydroxylation of histidinyl residues within ankyrin repeat domains - PubMed (original) (raw)
Factor-inhibiting hypoxia-inducible factor (FIH) catalyses the post-translational hydroxylation of histidinyl residues within ankyrin repeat domains
Ming Yang et al. FEBS J. 2011 Apr.
Free PMC article
Abstract
Factor-inhibiting hypoxia-inducible factor (FIH) is an Fe(II)/2-oxoglutarate-dependent dioxygenase that acts as a negative regulator of the hypoxia-inducible factor (HIF) by catalysing β-hydroxylation of an asparaginyl residue in its C-terminal transcriptional activation domain (CAD). In addition to the hypoxia-inducible factor C-terminal transcriptional activation domain (HIF-CAD), FIH also catalyses asparaginyl hydroxylation of many ankyrin repeat domain-containing proteins, revealing a broad sequence selectivity. However, there are few reports on the selectivity of FIH for the hydroxylation of specific residues. Here, we report that histidinyl residues within the ankyrin repeat domain of tankyrase-2 can be hydroxylated by FIH. NMR and crystallographic analyses show that the histidinyl hydroxylation occurs at the β-position. The results further expand the scope of FIH-catalysed hydroxylations.
© 2011 The Authors Journal compilation © 2011 FEBS.
Figures
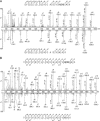
Figure 1
Tankryase-2 is a substrate for FIH-catalysed His hydroxylation. (A) Sequence alignment of the tankyrase-2 ARD (residues 57–798) [36] demonstrating a 4-repeat periodicity of the ARD, with insertion sequences (right) following boxed residues in the corresponding repeats to the left. Asn residues that have previously been identified as FIH substrates [8], and the two His residues located at the conserved hydroxylation position, are highlighted in bold and their residue number shown in parenthesis on the right. (B) Tankyrase-2 peptides containing His 238 and His 553 are FIH substrates in vitro. Peptides corresponding to: (I) TNKS2 223–243 (RVKIVQLLLQHGADVHAKDKG) and (II) TNKS2 538–558 (RVSVVEYLLQHGADVHAKDKG) were incubated in the presence of recombinant FIH and displayed a net +16 Da mass shift as determined by LC-MS. Subsequent MS/MS of the FIH-reacted TNKS2 538–558 peptide assigned the oxidation to His 553 (Fig. S1).
Figure 2
MS analyses assigning hydroxylation at His 238 and His 553 in tankyrase-2. Tankyrase-2 was purified from transiently transfected 293T cells coexpressing FIH. (A) MS/MS spectra of the tryptic peptide IVQLLLQHGADVHAK derived from tankyrase-2 (residues 226–240) in the hydroxylated ([M + 3H]3+ = m/z 553.28) (upper) and nonhydroxylated ([M + H]3+ = m/z 548.63) (lower) state. The hydroxylated species (upper) exhibits a + 16 Da mass shift on the y-ion series appearing at y3 and assigning hydroxylation to His238. (B) MS/MS of the tankyrase-2 tryptic peptide containing His 553 (VSVVEYLLQHGADVHAK) in hydroxylated ([M + 3H]3+ = m/z 627.64) (upper) and unmodified forms ([M + 3H]3+ = m/z 622.30) (lower). For both hydroxylated spectra, a −2 Da mass shift was commonly observed on fragment ions containing hydroxyhistidine, which is consistent with hydroxylation (+16 Da) followed by dehydration (−18 Da). Because there was no evidence for a −2 Da shift on the precursor ions it is likely that during the collision-induced dissociation process in the MS/MS analyses, the hydroxylated His residue undergoes dehydration to form the conjugated α,β-dehydrohistidine product. Note also there was no evidence for formation of the dehydrohistidine in the NMR analyses (Fig. 4).
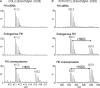
Figure 3
LC/MS spectra illustrating the effect of FIH intervention on tankyrase-2 hydroxylation at His 238 and His 553. 293T cells were treated with siRNAs against FIH (‘FIH siRNA’) or a control sequence (‘Endogenous FIH’/‘FIH overexpression’). After siRNA treatment, cells were transfected with FLAG–TNKS2 and either pcDNA3 FIH (‘FIH overexpression’) or empty vector (‘FIH siRNA’/‘Endogenous FIH’). FLAG–TNKS2 was immunopurified, digested and analysed by LC/MS to quantify hydroxylation. The efficacy of the siRNA and plasmid transfections were confirmed by anti-FIH and anti-FLAG immunoblotting (data not shown). (A) Representative LC/MS spectra of the 226–240 tryptic fragment containing His 238. Hydroxylation (∼ 30%) was observed for His 238 under conditions where the level of FIH was not limiting. (B) Representative LC/MS spectra of the 538–555 tryptic fragment containing His 553. Hydroxylation was observed at endogenous level of FIH (∼ 15%) and when FIH was overexpressed (∼ 70%).
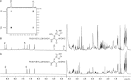
Figure 4
FIH catalysed His-hydroxylation occurs at the β-position. Hydroxylated TNKS2 538–558 peptide (RVSVVEYLLQHGADVHAKDKG) was produced by incubation with FIH under standard assay conditions (hydroxylated to ∼ 75% as assessed by MALDI-TOF analyses), LC-MS purified and analysed by NMR spectroscopy. (B) 1H NMR spectrum of the hydroxylated TNKS2 538–558 peptide in 2H2O. The resonances at 4.87 and 5.50 ppm, which are absent in the 1H NMR spectrum of the nonhydroxylated TNKS2 538–558 peptide (A), are ascribed to the α- and β-proton, respectively, of the hydroxylated His residues. The resonances for the imidazole ring protons (at positions 2 and 5) are deshielded in the spectrum of the hydroxylated peptide compared to that of the nonhydroxylated. (C) 2D 1H–1H COSY spectrum of the hydroxylated TNKS2 538–558 peptide in 2H2O indicating the 1H–1H correlation between resonances arising from the α- and β-hydrogens of the hydroxylated His residue.
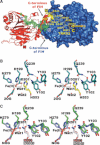
Figure 5
Structure of the FIH complexes. (A) Surface representation of the FIH·TNKS2 538–558 dimer structure (PDB ID: 2Y0I) to 2.28 Å resolution showing apparent electron density for residues Ser 540 to His 553 of the TNKS2 538–558 peptide (2_F_o − _F_c map, contoured to 1σ). (B) Stereoview stick representation of the FIH active site of the FIH·TNKS2 538–558 complex (FIH, deep teal; TNKS2 538–558, yellow; Fe(II), orange). (C) Stereoview stick representation of the superimposed FIH·mNotch1 1930–1949 (PDB ID: 3P3N, FIH in green and N1 1930–1949 in white) and FIH·HIF-1αCAD 788–826 (PDB ID: 1H2K; FIH in purple and HIF-1αCAD 788-826 in salmon) complexes. A comparison of all FIH complexes suggests that the pro-S hydrogen of His 553 in TNKS2 538–558 is likely analogously positioned as that observed for hydroxylated asparagines in FIH·mNotch1 1930–1949 (PDB ID: 3P3N) and FIH·HIF-1αCAD 788–826 (PDB ID: 1H2K) complexes. (B) and (C) also illustrate the differences in side-chain conformation for Gln 239FIH and Tyr 103FIH between the FIH·TNKS2 538–558 and FIH·mNotch1 1930–1949/FIH·HIF-1αCAD 788-826 complexes.
Figure 6
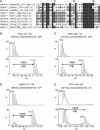
FIH-catalysed His hydroxylation of ARDs may be common. (A)
clustalw
nongapped multiple sequence alignment of ankyrin repeat sequences containing a target histidine residue at the conserved hydroxylation position. Corresponding peptides spanning the potential histidinyl hydroxylation sites were tested as FIH substrates in vitro, among which peptides derived from TNKS1, GABPB2 and TRPV4 demonstrate FIH-dependent hydroxylation. (B–D) LC/MS analyses demonstrating FIH-catalysed His hydroxylation of the following peptides: (B) TNKS1 381–400; (C) TNKS1 696–715; and (D) GABPB2 115–135. (E) MALDI spectra showing the hydroxylation of the TRPV4 249–269 peptide.
Similar articles
- Proteomics-based identification of novel factor inhibiting hypoxia-inducible factor (FIH) substrates indicates widespread asparaginyl hydroxylation of ankyrin repeat domain-containing proteins.
Cockman ME, Webb JD, Kramer HB, Kessler BM, Ratcliffe PJ. Cockman ME, et al. Mol Cell Proteomics. 2009 Mar;8(3):535-46. doi: 10.1074/mcp.M800340-MCP200. Epub 2008 Oct 20. Mol Cell Proteomics. 2009. PMID: 18936059 Free PMC article. - Posttranslational hydroxylation of ankyrin repeats in IkappaB proteins by the hypoxia-inducible factor (HIF) asparaginyl hydroxylase, factor inhibiting HIF (FIH).
Cockman ME, Lancaster DE, Stolze IP, Hewitson KS, McDonough MA, Coleman ML, Coles CH, Yu X, Hay RT, Ley SC, Pugh CW, Oldham NJ, Masson N, Schofield CJ, Ratcliffe PJ. Cockman ME, et al. Proc Natl Acad Sci U S A. 2006 Oct 3;103(40):14767-72. doi: 10.1073/pnas.0606877103. Epub 2006 Sep 26. Proc Natl Acad Sci U S A. 2006. PMID: 17003112 Free PMC article. - MYPT1, the targeting subunit of smooth-muscle myosin phosphatase, is a substrate for the asparaginyl hydroxylase factor inhibiting hypoxia-inducible factor (FIH).
Webb JD, Murányi A, Pugh CW, Ratcliffe PJ, Coleman ML. Webb JD, et al. Biochem J. 2009 May 13;420(2):327-33. doi: 10.1042/BJ20081905. Biochem J. 2009. PMID: 19245366 - FIH-dependent asparaginyl hydroxylation of ankyrin repeat domain-containing proteins.
Cockman ME, Webb JD, Ratcliffe PJ. Cockman ME, et al. Ann N Y Acad Sci. 2009 Oct;1177:9-18. doi: 10.1111/j.1749-6632.2009.05042.x. Ann N Y Acad Sci. 2009. PMID: 19845602 Review. - Factor inhibiting hypoxia-inducible factor (FIH) and other asparaginyl hydroxylases.
Lancaster DE, McDonough MA, Schofield CJ. Lancaster DE, et al. Biochem Soc Trans. 2004 Dec;32(Pt 6):943-5. doi: 10.1042/BST0320943. Biochem Soc Trans. 2004. PMID: 15506931 Review.
Cited by
- Oxygen-regulated post-translation modifications as master signalling pathway in cells.
Batie M, Fasanya T, Kenneth NS, Rocha S. Batie M, et al. EMBO Rep. 2023 Dec 6;24(12):e57849. doi: 10.15252/embr.202357849. Epub 2023 Oct 25. EMBO Rep. 2023. PMID: 37877678 Free PMC article. Review. - The Jumonji-C oxygenase JMJD7 catalyzes (3S)-lysyl hydroxylation of TRAFAC GTPases.
Markolovic S, Zhuang Q, Wilkins SE, Eaton CD, Abboud MI, Katz MJ, McNeil HE, Leśniak RK, Hall C, Struwe WB, Konietzny R, Davis S, Yang M, Ge W, Benesch JLP, Kessler BM, Ratcliffe PJ, Cockman ME, Fischer R, Wappner P, Chowdhury R, Coleman ML, Schofield CJ. Markolovic S, et al. Nat Chem Biol. 2018 Jul;14(7):688-695. doi: 10.1038/s41589-018-0071-y. Epub 2018 Jun 18. Nat Chem Biol. 2018. PMID: 29915238 Free PMC article. - Kinetic Investigations of the Role of Factor Inhibiting Hypoxia-inducible Factor (FIH) as an Oxygen Sensor.
Tarhonskaya H, Hardy AP, Howe EA, Loik ND, Kramer HB, McCullagh JS, Schofield CJ, Flashman E. Tarhonskaya H, et al. J Biol Chem. 2015 Aug 7;290(32):19726-42. doi: 10.1074/jbc.M115.653014. Epub 2015 Jun 25. J Biol Chem. 2015. PMID: 26112411 Free PMC article. - Factor inhibiting HIF (FIH) recognizes distinct molecular features within hypoxia-inducible factor-α (HIF-α) versus ankyrin repeat substrates.
Wilkins SE, Karttunen S, Hampton-Smith RJ, Murchland I, Chapman-Smith A, Peet DJ. Wilkins SE, et al. J Biol Chem. 2012 Mar 16;287(12):8769-81. doi: 10.1074/jbc.M111.294678. Epub 2012 Jan 23. J Biol Chem. 2012. PMID: 22270367 Free PMC article. - Selective Hypoxia-Sensitive Oxomer Formation by FIH Prevents Binding of the NF-κB Inhibitor IκBβ to NF-κB Subunits.
Volkova YL, Jucht AE, Oechsler N, Krishnankutty R, von Kriegsheim A, Wenger RH, Scholz CC. Volkova YL, et al. Mol Cell Biol. 2024;44(4):138-148. doi: 10.1080/10985549.2024.2338727. Epub 2024 Apr 22. Mol Cell Biol. 2024. PMID: 38644795 Free PMC article.
References
- Hewitson KS, McNeill LA, Riordan MV, Tian YM, Bullock AN, Welford RW, Elkins JM, Oldham NJ, Bhattacharya S, Gleadle JM, et al. Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J Biol Chem. 2002;277:26351–26355. - PubMed
- Koivunen P, Hirsila M, Gunzler V, Kivirikko KI, Myllyharju J. Catalytic properties of the asparaginyl hydroxylase (FIH) in the oxygen sensing pathway are distinct from those of its prolyl 4-hydroxylases. J Biol Chem. 2004;279:9899–9904. - PubMed
- Cockman ME, Lancaster DE, Stolze IP, Hewitson KS, McDonough MA, Coleman ML, Coles CH, Yu X, Hay RT, Ley SC, et al. Posttranslational hydroxylation of ankyrin repeats in IkappaB proteins by the hypoxia-inducible factor (HIF) asparaginyl hydroxylase, factor inhibiting HIF (FIH) Proc Natl Acad Sci USA. 2006;103:14767–14772. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous