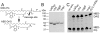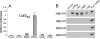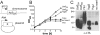Enzymatic activities and functional interdependencies of Bacillus subtilis lipoteichoic acid synthesis enzymes - PubMed (original) (raw)
Enzymatic activities and functional interdependencies of Bacillus subtilis lipoteichoic acid synthesis enzymes
Mirka E Wörmann et al. Mol Microbiol. 2011 Feb.
Abstract
Lipoteichoic acid (LTA) is an important cell wall polymer in Gram-positive bacteria. The enzyme responsible for polyglycerolphosphate LTA synthesis is LtaS, first described in Staphylococcus aureus. Four LtaS orthologues, LtaS(BS) , YfnI, YqgS and YvgJ, are present in Bacillus subtilis. Using an in vitro enzyme assay, we determined that all four proteins are Mn(2+) -dependent metal enzymes that use phosphatidylglycerol as a substrate. We show that LtaS(BS) , YfnI and YqgS can produce polymers, suggesting that these three proteins are bona-fide LTA synthases while YvgJ functions as an LTA primase, as indicated by the accumulation of a GroP-Glc(2) -DAG glycolipid. Western blot analysis of LTA produced by ltaS(BS) , yfnI, yqgS and yvgJ single, triple and the quadruple mutant, showed that LTA production was only abolished in the quadruple and the YvgJ-only expressing mutant. B. subtilis strains expressing YfnI in the absence of LtaS(BS) produced LTA of retarded mobility, presumably caused by an increase in chain length as suggested by a structural analysis of purified LTA. Taken together, the presented results indicate that the mere presence or absence of LTA cannot account for cell division and sporulation defects observed in the absence of individual enzymes and revealed an unexpected enzymatic interdependency of LtaS-type proteins in B. subtilis.
© 2010 Blackwell Publishing Ltd.
Figures
Fig. 1
In vitro activity of B. subtilis LtaS-type enzymes. A. Chemical structures of fluorescently labelled NBD-PG and NBD-DAG lipids with known S. aureus LtaS and B. cereus PLC cleavage site indicated by an arrow. B. Coomassie stained gel of purified B. subtilis LtaS-like proteins. Extracellular enzymatic domains of B. subtilis LtaSBS, YfnI, YqgS and YvgJ were purified as N-terminal His-tag fusion proteins and 10 µg purified protein separated on a 10% SDS-PAGE gel and visualized by staining with Coomassie brilliant blue. C. TLC analysis of B. subtilis LtaSBS, YfnI, YvgJ and YqgS in vitro reaction products. The NBD-PG lipid substrate was incubated with eLtaSBS, eYfnI, eYvgJ or eYqgS enzyme. Subsequently, lipids were extracted and separated by TLC and fluorescent lipid bands visualized by scanning plates with a fluorescence imager. As negative and positive controls, reactions were set up without enzyme or with the B. cereus PLC enzyme respectively. Note that only 10% of the PLC reaction was run on the TLC plate. Positions of NBD-PG and presumed NBD-DAG reaction product are indicated on the left and proteins added to each reaction are shown on the top of the panel.
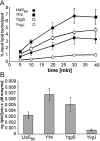
Fig. 2
Kinetic measurements for recombinant LtaSBS, YfnI, YqgS and YvgJ enzymes. A. Time-course experiment. Enzyme reactions were set up as described under Experimental procedures, aliquots removed at the indicated time points and reactions stopped by the addition of chloroform and methanol. Lipids were separated on TLC plates and the NBD-DAG reaction product quantified. For each time point and enzyme the average value and standard deviation of three values is plotted. Three independent experiments were performed and a representative graph is shown. B. Maximal enzyme activity of B. subtilis LtaSBS, YfnI, YqgS and YvgJ. The slope of the linear fit through the first three data points of the curve shown in (A) was used to calculate the maximal enzyme activity for each B. subtilis LtaS orthologue. Three independent time-course experiments were used to determine an average value and standard deviation for the maximal enzyme activity and these values are plotted.
Fig. 3
Metal and substrate specificity of recombinant B. subtilis LtaS-type enzymes. A. B. subtilis LtaS-type enzymes require Mn2+ for activity. In vitro enzyme assays were set up with NBD-PG lipid as the substrate in the presence of 10 mM MgCl2, MnCl2, CaCl2 or ZnCl2 and reactions were initiated by the addition of eLtaSBS. As controls, reactions were set up without enzyme or without metal ion added. Samples were incubated for 3 h at 37°C, lipids extracted and separated by TLC. Plates were scanned and signals of the reaction product quantified. Reactions were set up in triplicate and the average value and standard deviation plotted. The average fluorescence reading for the reactions set up with MnCl2 was set to 1 and other values were adjusted accordingly. Similar results were obtained for B. subtilis YfnI, YqgS and YvgJ (see Fig. S2). B. NBD-PG is the sole lipid substrate for B. subtilis LtaSBS, YfnI, YqgS and YvgJ. Standard enzyme reactions were set up using NBD-PG, NBD-PS, NBD-PE or NBD-PC as substrate (indicated on the left of the panel) and reactions were initiated by the addition of the different B. subtilis enzymes. As a negative control, lipid substrates were incubated without enzyme (no enz.) and as a positive control, a PLC reaction using NBD-PG as substrate was run alongside on each TLC plate in order to determine the mobility of the reaction product. Three independent experiments were performed and a representative result is shown. Note that only the upper part of the TLC plates is shown with the area of the reaction product.
Fig. 4
Functional complementation of an _S. aureus ltaS_-depletion strain with B. subtilis ltaSBS, yfnI, yqgS or yvgJ expressed from a multicopy plasmid. A. Schematic representation of complementation strains. S. aureus strains used for complementation analysis contain the chromosomal copy of ltaS under IPTG inducible expression control and harbour a multicopy plasmid (pCN34) for expression of LtaS orthologues from the tetracycline inducible promoter. B. Bacterial growth curves. Washed overnight cultures of S. aureus strains ANG1571 (LtaSSA-expressing), ANG1662 (LtaSBS-expressing), ANG1573 (YfnI-expressing), ANG1654 (YqgS-expressing) (ANG1658) (YvgJ-expression) and ANG1130 (containing empty vector pCN34; no insert as negative control) were diluted 1:100 into fresh medium containing 300 ng ml−1 Atet and growth was monitored by determining OD600 readings at the indicated time points. All cultures were back-diluted 1:100 at the 4 h time point and cultures with strains expressing LtaSSA, LtaSBS and YqgS were back diluted a second time at the 8 h time point to maintain cultures in the logarithmic growth phase. C. LTA detection by Western blot. The same S. aureus strains and growth conditions as described above were used for LTA analysis by Western blot. At the 4 h time point, 1 ml culture aliquots were removed and samples prepared and analysed by Western blot as described in the Experimental procedures section. For LTA detection, the mouse monoclonal LTA antibody (Clone 55 from Hycult biotechnology) and the HRP-conjugated anti-mouse IgG antibody (Cell Signaling Technologies, USA) were used at 1:5000 and 1:10 000 dilutions respectively. Sizes of proteins standards run in parallel are shown on the left of the panel and proteins expressed in each strain are given above each lane. Note that the LTA Western blot with samples isolated from S. aureus strains expressing YqgS, YvgJ or containing the empty vector (no insert) was exposed four times longer.
Fig. 5
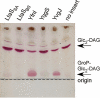
TLC analysis of glycolipids. S. aureus strains ANG1571 (LtaSSA-expressing), ANG1662 (LtaSBS-expressing), ANG1573 (YfnI-expressing), ANG1654 (YqgS-expressing), ANG1658 (YvgJ-expressing) and ANG1130 (containing empty vector pCN34, no insert as negative control) were grown to mid-log phase and lipids extracted as described in the Experimental procedures section. Five hundred µg total membrane lipids were separated by TLC and glycolipids visualized by staining with α-naphthol/sulphuric acid. The position of the origin is indicated by a dashed line, positions of presumed Glc2-DAG (top band) and GroP-Glc2-DAG (bottom band) lipids are marked with arrows on the right of the panel and proteins expressed in the different strains are indicated above each lane.
Fig. 6
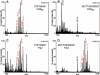
MALDI-TOF analysis of glycolipids produced by LtaSBS and YvgJ-expressing S. aureus strains. A total of 2.5 mg lipids isolated from LtaSBS or YvgJ-expressing S. aureus strains were separated by TLC and lipids corresponding to top and bottom glycolipids bands extracted and analysed by MALDI-TOF mass spectrometry. Spectra were recorded in the reflector positive ion mode and are shown for (A) LtaSBS top band, (B) LtaSBS bottom band, (C) YvgJ top band and (D) YvgJ bottom band. Maximal signal intensity is shown in the top right corner in each panel. Note that the maximum signal intensity in panel B is lower than in the other three panels, which amplifies background signals in the normalized representation shown. Observed masses corresponding to calculated masses of glycolipids are shown in red. Three independent experiments were performed and representative spectra are shown.
Fig. 7
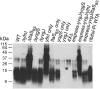
LTA production by wild-type and mutant B. subtilis strains. Samples for LTA analysis by Western blot were prepared from overnight cultures of wild-type and mutant B. subtilis 168 strains and from a B. subtilis 168 hybrid strain expressing ribitolphosphate wall teichoic acid. Samples were separated on a 15% SDS-PAGE gel, transferred to a PVDF membrane and LTA detected by Western blot using the humanized monoclonal LTA-specific antibody (Biosynexus Incorporated) and the HRP-linked anti-human antibody (DakoCytomation) at 1:10 000 dilutions. Sizes of protein standards run in parallel are indicated on the left of the panel and strains used are indicated above each lane, with abbreviations given in strain Table 2.
Fig. 8
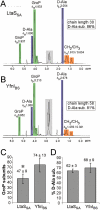
NMR and biochemical analysis of purified LTA. A and B. NMR analysis. Large cultures of S. aureus strains (A) ANG514 (LtaSSA-expressing) and (B) ANG515 (YfnI-expressing) were grown and LTA purified as described in the Experimental procedures section. One milligram purified LTA was suspended and lyophilized several times in D2O to exchange 1H for 2H deuterons and 1H NMR spectra were recorded at 600 MHz, 300 K. The signals derived from citrate, a buffer component used during LTA purification and retained in the samples are marked in grey. The different signals previously assigned to LTA components (Morath et al., 2002) are colour coded [blue – D-Ala (4 protons per D-Ala group), green – GroP (5 protons per GroP group), orange – CH2/CH3 groups of fatty acids (59 protons per lipid anchor)]. The integration values are shown above each signal. Chain length was determined by calculating the ratio of integral values for GroP to CH2/CH3 groups in fatty acids and % D-Ala substitution by calculating the ratio of integral values for D-Ala to GroP x 100 and taking into account the number of protons for each signal. NMR analysis was performed on four independently isolated LTA samples for each strain and a representative result is shown. C and D. Biochemical analysis of LTA. LTA extracted from strains ANG514 (LtaSSA) and ANG515 (YfnIBS) was subjected to a biochemical analysis. Phosphate, glucose and D-Ala contents were determined as described in the Experimental procedures section. GroP, D-Ala and glucose solutions of known concentrations were used as standards. The chain length in GroP subunits (C) was determined by calculation of the ratio of phosphate/½ glucose concentration and the % D-Ala substitution (D) by calculating the ratio of D-Ala/phosphate concentration × 100. Biochemical analysis was performed on four independently isolated LTA samples for each strain and the mean and standard deviation is shown. The difference in chain length is statistically significant and indicated with an asterisk (*) (two-tailed _P_-value of 0.017; unpaired _T_-test) while the difference in D-Ala modification is not (two-tailed _P_-value of 0.355, unpaired _T_-test).
Fig. 9
Schematic representation of in vivo activities of the four B. subtilis LtaS-type enzymes. A. B. subtilis LtaSBS is the ‘house-keeping’ LTA synthase, which is active during vegetative growth. B. B. subtilis YfnI is assumed to be the ‘stress’ LTA synthase as yfnI transcription is controlled by sigma M, which is important during cell envelope stress. YfnI is capable of promoting polyglycerolphosphate synthesis as well as producing the GroP-Glc2-DAG glycolipid intermediate. Here we show that YfnI activity is influenced by the presence/absence of LtaSBS. Processed forms of both, LtaSBS and YfnI have been detected in the culture supernatant and processing of YfnI is reduced in the combined absence of the two signal peptidases SipT and SipV (Antelmann et al., 2001). C. B. subtilis YqgS has LTA synthase activity and is important during the sporulation process.D. YvgJ functions as an LTA primase synthesizing the glycolipid intermediate GroP-Glc2-DAG. Although YqgS and YvgJ contain an AXA motif it is not clear if these enzymes are processed in B. subtilis. LTA synthases are depicted in blue and LTA primases in red. Numbers refer to amino acid positions and arrows indicate cleavage or potential cleavage sites.
Similar articles
- Induction of extracytoplasmic function sigma factors in Bacillus subtilis cells with defects in lipoteichoic acid synthesis.
Hashimoto M, Seki T, Matsuoka S, Hara H, Asai K, Sadaie Y, Matsumoto K. Hashimoto M, et al. Microbiology (Reading). 2013 Jan;159(Pt 1):23-35. doi: 10.1099/mic.0.063420-0. Epub 2012 Oct 25. Microbiology (Reading). 2013. PMID: 23103977 - Localization and expression of the Bacillus subtilis DL-endopeptidase LytF are influenced by mutations in LTA synthases and glycolipid anchor synthetic enzymes.
Kiriyama Y, Yazawa K, Tanaka T, Yoshikawa R, Yamane H, Hashimoto M, Sekiguchi J, Yamamoto H. Kiriyama Y, et al. Microbiology (Reading). 2014 Dec;160(Pt 12):2639-2649. doi: 10.1099/mic.0.080366-0. Epub 2014 Oct 6. Microbiology (Reading). 2014. PMID: 25288647 - Synthesis of glycerol phosphate lipoteichoic acid in Staphylococcus aureus.
Gründling A, Schneewind O. Gründling A, et al. Proc Natl Acad Sci U S A. 2007 May 15;104(20):8478-83. doi: 10.1073/pnas.0701821104. Epub 2007 May 3. Proc Natl Acad Sci U S A. 2007. PMID: 17483484 Free PMC article. - Location, synthesis and function of glycolipids and polyglycerolphosphate lipoteichoic acid in Gram-positive bacteria of the phylum Firmicutes.
Reichmann NT, Gründling A. Reichmann NT, et al. FEMS Microbiol Lett. 2011 Jun;319(2):97-105. doi: 10.1111/j.1574-6968.2011.02260.x. Epub 2011 Mar 25. FEMS Microbiol Lett. 2011. PMID: 21388439 Free PMC article. Review. - Lipoteichoic acid synthesis and function in gram-positive bacteria.
Percy MG, Gründling A. Percy MG, et al. Annu Rev Microbiol. 2014;68:81-100. doi: 10.1146/annurev-micro-091213-112949. Epub 2014 May 5. Annu Rev Microbiol. 2014. PMID: 24819367 Review.
Cited by
- Magnesium rescues the morphology of Bacillus subtilis mreB mutants through its inhibitory effect on peptidoglycan hydrolases.
Tesson B, Dajkovic A, Keary R, Marlière C, Dupont-Gillain CC, Carballido-López R. Tesson B, et al. Sci Rep. 2022 Jan 21;12(1):1137. doi: 10.1038/s41598-021-04294-5. Sci Rep. 2022. PMID: 35064120 Free PMC article. - Metabolism Shapes the Cell.
Sperber AM, Herman JK. Sperber AM, et al. J Bacteriol. 2017 May 9;199(11):e00039-17. doi: 10.1128/JB.00039-17. Print 2017 Jun 1. J Bacteriol. 2017. PMID: 28320879 Free PMC article. Review. - c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress.
Corrigan RM, Abbott JC, Burhenne H, Kaever V, Gründling A. Corrigan RM, et al. PLoS Pathog. 2011 Sep;7(9):e1002217. doi: 10.1371/journal.ppat.1002217. Epub 2011 Sep 1. PLoS Pathog. 2011. PMID: 21909268 Free PMC article. - TerC proteins function during protein secretion to metalate exoenzymes.
He B, Sachla AJ, Helmann JD. He B, et al. Nat Commun. 2023 Oct 4;14(1):6186. doi: 10.1038/s41467-023-41896-1. Nat Commun. 2023. PMID: 37794032 Free PMC article. - Extraction and Analysis of Bacterial Teichoic Acids.
Kho K, Meredith TC. Kho K, et al. Bio Protoc. 2018 Nov 5;8(21):e3078. doi: 10.21769/BioProtoc.3078. eCollection 2018 Nov 5. Bio Protoc. 2018. PMID: 34532535 Free PMC article.
References
- Antelmann H, Tjalsma H, Voigt B, Ohlmeier S, Bron S, van Dijl JM, Hecker M. A proteomic view on genome-based signal peptide predictions. Genome Res. 2001;11:1484–1502. - PubMed
- Burkholder PR, Giles NH. Induced biochemical mutations in Bacillus subtilis. Am J Bot. 1947;33:345–348. - PubMed
- Chiu TH, Morimoto H, Baker JJ. Biosynthesis and characterization of phosphatidylglycerophosphoglycerol, a possible intermediate in lipoteichoic acid biosynthesis in Streptococcus sanguis. Biochim Biophys Acta. 1993;1166:222–228. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases