Genomic signatures of fifth autotrophic carbon assimilation pathway in bathypelagic Crenarchaeota - PubMed (original) (raw)
Genomic signatures of fifth autotrophic carbon assimilation pathway in bathypelagic Crenarchaeota
Violetta La Cono et al. Microb Biotechnol. 2010 Sep.
Abstract
Marine Crenarchaeota, ubiquitous and abundant organisms in the oceans worldwide, remain metabolically uncharacterized, largely due to their low cultivability. Identification of candidate genes for bicarbonate fixation pathway in the Cenarchaeum symbiosum A was an initial step in understanding the physiology and ecology of marine Crenarchaeota. Recent cultivation and genome sequencing of obligate chemoautotrophic Nitrosopumilus maritimus SCM1 were a major breakthrough towards understanding of their functioning and provide a valuable model for experimental validation of genomic data. Here we present the identification of multiple key components of 3-hydroxipropionate/4-hydroxybutyrate cycle, the fifth pathway in carbon fixation, found in data sets of environmental sequences representing uncultivated superficial and bathypelagic Crenarchaeota from Sargasso sea (GOS data set) and KM3 (Mediterranean Sea) and ALOHA (Atlantic ocean) stations. These organisms are likely to use acetyl-CoA/propionyl-CoA carboxylase(s) as CO₂-fixing enzyme(s) to form succinyl-CoA, from which one molecule of acetyl-CoA is regenerated via 4-hydroxybutyrate cleavage and another acetyl-CoA to be the pathway product. The genetic distinctiveness and matching sympatric abundance imply that marine crenarchaeal genotypes from the three different geographic sites share similar ecophysiological properties, and therefore may represent fundamental units of marine ecosystem functioning. To couple results of sequence comparison with the dark ocean primary production, dissolved inorganic carbon fixation rates were measured at KM3 Station (3000 m depth, Eastern Mediterranean Sea), i.e. at the same site and depth used for metagenomic library construction.
© 2010 The Authors. Journal compilation © 2010 Society for Applied Microbiology and Blackwell Publishing Ltd.
Figures
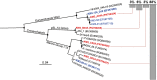
Figure 1
Phylogenetic tree of archaeal 16S rRNA genes amplified from the pool of environmental DNA recovered from 3010 m at KM3 Station. The tree was constructed by Neighbour‐Joining method and Jukes–Cantor distance matrix using MacVector 11.0.2 and a total of 550 non‐ambiguously aligned positions. Non‐parametric bootstrapping was performed upon 1000 replicates. Bootstrap values > 70 and ≥ 80 are shown as empty and filled circles respectively. The tree was rooted with Alteromonas genoviensis strain I96 16S ribosomal RNA gene (FJ040187). KM3 water column sequences obtained in this work and previously (Martín‐Cuadrado et al., 2007) are shown in red and blue respectively. Additionally, the 16S rDNA sequences found in KM3 fosmid library are shown in bold.
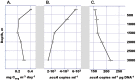
Figure 2
Prokaryotic dark ocean primary production rates (A); quantitative PCR‐determined gene copy numbers of crenarchaeal accA at different depths of KM3 water column given as absolute (B) and normalized against the total DNA (C). All error bars indicate standard deviation from a mean of three replicate measurements.
Figure 3
Comparison of genomic fragments containing acetyl(propionyl)‐CoA carboxylase (accA) operon in N. maritimus and C. symbiosum with five related environmental fosmid sequences, obtained from superficial (GOS database, red) and bathypelagic seawater (4000 m WGS ALOHA database, blue). A. Genes shared in common between genomic fragments are connected by: red (acc operon), blue (nuo operon) and grey (rest of the genes) shaded boxes. B. Re‐assembling of individual reads in HF4000_ANIW97P9 and HF4000_APKG6D3 is depicted in grey insert. C. Genomic fragments shared significant conservation of gene content and order in all analysed sequences.
Figure 4
Comparison of the genomic fragments containing methylmalonyl‐CoA mutase/epimerase operon in N. maritimus and C. symbiosum with three related environmental fosmid sequences, obtained from superficial (GOS database, red) and bathypelagic seawater (ALOHA and KM3 database, blue). A. Genes shared by genomic fragments are connected by shaded boxes: red (3‐HP/4‐HB cycle), blue (triosephosphate formation pathway) and grey (rest of the genes). B. The core genomic fragment of 19 kb shared significant conservation in gene content and order in all analysed sequences.
Figure 5
Comparison of genomic fragments harbouring 4‐hydroxybutyryl‐CoA dehydratase in N. maritimus and C. symbiosum with four related environmental fosmid sequences, obtained from superficial (GOS database, red) and bathypelagic seawater (4000 m WGS ALOHA database, blue). A. Genes shared in common between genomic fragments are connected by shaded boxes: red (3‐HP/4‐HB cycle), blue (triosephosphate formation and glyoxylate pathway) and grey (rest of the genes). B. Re‐assemblage of individual reads in HF4000_APKG7F11 and HF4000_ANIW133M9 is depicted in grey insert. C. The genomic fragments share significant conservation of gene content and order in all analysed sequences.
Similar articles
- Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota.
Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM, DeLong EF. Hallam SJ, et al. PLoS Biol. 2006 Apr;4(4):e95. doi: 10.1371/journal.pbio.0040095. Epub 2006 Mar 21. PLoS Biol. 2006. PMID: 16533068 Free PMC article. - Contribution of crenarchaeal autotrophic ammonia oxidizers to the dark primary production in Tyrrhenian deep waters (Central Mediterranean Sea).
Yakimov MM, Cono VL, Smedile F, DeLuca TH, Juárez S, Ciordia S, Fernández M, Albar JP, Ferrer M, Golyshin PN, Giuliano L. Yakimov MM, et al. ISME J. 2011 Jun;5(6):945-61. doi: 10.1038/ismej.2010.197. Epub 2011 Jan 6. ISME J. 2011. PMID: 21209665 Free PMC article. - Convergent Evolution of a Promiscuous 3-Hydroxypropionyl-CoA Dehydratase/Crotonyl-CoA Hydratase in Crenarchaeota and Thaumarchaeota.
Liu L, Brown PC, Könneke M, Huber H, König S, Berg IA. Liu L, et al. mSphere. 2021 Jan 20;6(1):e01079-20. doi: 10.1128/mSphere.01079-20. mSphere. 2021. PMID: 33472982 Free PMC article. - Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota.
Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Brochier-Armanet C, et al. Nat Rev Microbiol. 2008 Mar;6(3):245-52. doi: 10.1038/nrmicro1852. Nat Rev Microbiol. 2008. PMID: 18274537 Review. - Molecular signatures for the Crenarchaeota and the Thaumarchaeota.
Gupta RS, Shami A. Gupta RS, et al. Antonie Van Leeuwenhoek. 2011 Feb;99(2):133-57. doi: 10.1007/s10482-010-9488-3. Epub 2010 Aug 14. Antonie Van Leeuwenhoek. 2011. PMID: 20711675 Review.
Cited by
- The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology.
Pester M, Schleper C, Wagner M. Pester M, et al. Curr Opin Microbiol. 2011 Jun;14(3):300-6. doi: 10.1016/j.mib.2011.04.007. Epub 2011 May 4. Curr Opin Microbiol. 2011. PMID: 21546306 Free PMC article. Review. - Contribution of Bicarbonate Assimilation to Carbon Pool Dynamics in the Deep Mediterranean Sea and Cultivation of Actively Nitrifying and CO2-Fixing Bathypelagic Prokaryotic Consortia.
La Cono V, Ruggeri G, Azzaro M, Crisafi F, Decembrini F, Denaro R, La Spada G, Maimone G, Monticelli LS, Smedile F, Giuliano L, Yakimov MM. La Cono V, et al. Front Microbiol. 2018 Jan 19;9:3. doi: 10.3389/fmicb.2018.00003. eCollection 2018. Front Microbiol. 2018. PMID: 29403458 Free PMC article. - Community structure and function of planktonic Crenarchaeota: changes with depth in the South China Sea.
Hu A, Jiao N, Zhang CL. Hu A, et al. Microb Ecol. 2011 Oct;62(3):549-63. doi: 10.1007/s00248-011-9866-z. Epub 2011 May 20. Microb Ecol. 2011. PMID: 21597940 - Identification of missing genes and enzymes for autotrophic carbon fixation in crenarchaeota.
Ramos-Vera WH, Weiss M, Strittmatter E, Kockelkorn D, Fuchs G. Ramos-Vera WH, et al. J Bacteriol. 2011 Mar;193(5):1201-11. doi: 10.1128/JB.01156-10. Epub 2010 Dec 17. J Bacteriol. 2011. PMID: 21169482 Free PMC article. - Editorial - preview.
Giuliano L, Barbier M, Briand F. Giuliano L, et al. Microb Biotechnol. 2010 Sep;3(5):489-90. doi: 10.1111/j.1751-7915.2010.00206.x. Microb Biotechnol. 2010. PMID: 21255347 Free PMC article. No abstract available.
References
- Agogué H., Brink M., Dinasquet J., Herndl G.J. Major gradients in putatively nitrifying and non‐nitrifying Archaea in the deep North Atlantic. Nature. 2008;456:788–791. - PubMed
- Berg I.A., Kockelkorn D., Buckel W., Fuchs G. A 3‐hydroxypropionate/4‐hydrdoxybutyrate autotrophic carbon dixide assimilation pathway in Archaea. Science. 2007;318:1782–1786. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases