Microbial community composition and function in permanently cold seawater and sediments from an arctic fjord of svalbard - PubMed (original) (raw)
Microbial community composition and function in permanently cold seawater and sediments from an arctic fjord of svalbard
A Teske et al. Appl Environ Microbiol. 2011 Mar.
Abstract
Heterotrophic microbial communities in seawater and sediments metabolize much of the organic carbon produced in the ocean. Although carbon cycling and preservation depend critically on the capabilities of these microbial communities, their compositions and capabilities have seldom been examined simultaneously at the same site. To compare the abilities of seawater and sedimentary microbial communities to initiate organic matter degradation, we measured the extracellular enzymatic hydrolysis rates of 10 substrates (polysaccharides and algal extracts) in surface seawater and bottom water as well as in surface and anoxic sediments of an Arctic fjord. Patterns of enzyme activities differed between seawater and sediments, not just quantitatively, in accordance with higher cell numbers in sediments, but also in their more diversified enzyme spectrum. Sedimentary microbial communities hydrolyzed all of the fluorescently labeled polysaccharide and algal extracts, in most cases at higher rates in subsurface than surface sediments. In seawater, in contrast, only 5 of the 7 polysaccharides and 2 of the 3 algal extracts were hydrolyzed, and hydrolysis rates in surface and deepwater were virtually identical. To compare bacterial communities, 16S rRNA gene clone libraries were constructed from the same seawater and sediment samples; they diverged strongly in composition. Thus, the broader enzymatic capabilities of the sedimentary microbial communities may result from the compositional differences between seawater and sedimentary microbial communities, rather than from gene expression differences among compositionally similar communities. The greater number of phylum- and subphylum-level lineages and operational taxonomic units in sediments than in seawater samples may reflect the necessity of a wider range of enzymatic capabilities and strategies to access organic matter that has already been degraded during passage through the water column. When transformations of marine organic matter are considered, differences in community composition and their different abilities to access organic matter should be taken into account.
Figures
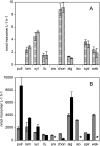
FIG. 1.
Hydrolysis rates of polysaccharides and plankton extracts in surface-water (horizontal-stripe bars) and bottom-water (slanted-stripe bars) samples (A) and in homogenized sediments from depths of 0 to 2 cm (gray bars) and 3 to 9 cm (black bars) (B). Substrates are pull, pullulan; lam, laminarin; xyl, xylan; fu, fucoidan; ara, arabinogalactan; chon, chondroitin sulfate; alg, alginic acid; iso, Isochrysis extract; spir, Spirulina extract; wak, wakame extract. Error bars show standard deviations of triplicate incubations. An asterisk indicates no data.
FIG. 2.
Percent representation of bacterial phylum- and subphylum-level groups in 16S rRNA gene clone libraries from water column and sediment samples in Smeerenburg Fjord, Svalbard. From left to right, surface water, deep water, surface sediment, and deep sediment community. cmbsf, centimeters below the surface.
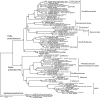
FIG. 3.
Neighbor-joining phylogeny of Smeerenburg Fjord alpha- and deltaproteobacterial phylotypes, based on an ∼1,200-bp alignment of bacterial 16S rRNA sequences. The Svalbard phylotypes are labeled with the habitat indicator (water_surface, water_deep, sediment_surface, sediment_deep), followed by the clone number, and are highlighted in boldface.
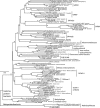
FIG. 4.
Neighbor-joining phylogeny of Smeerenburg Fjord Bacteroides phylotypes, based on an ∼1,200-bp alignment of bacterial 16S rRNA sequences. The Svalbard phylotypes are labeled with the habitat indicator (water_surface, water_deep, sediment_surface, sediment_deep), followed by the clone number, and are highlighted in boldface.
FIG. 5.
Neighbor-joining phylogeny of Smeerenburg Fjord beta- and gammaproteobacterial phylotypes, based on an ∼1,200-bp alignment of bacterial 16S rRNA sequences. The Svalbard phylotypes are labeled with the habitat indicator (water_surface, water_deep, sediment_surface, sediment_deep), followed by the clone number, and are highlighted in boldface.
Similar articles
- Functional differences between Arctic seawater and sedimentary microbial communities: contrasts in microbial hydrolysis of complex substrates.
Arnosti C. Arnosti C. FEMS Microbiol Ecol. 2008 Nov;66(2):343-51. doi: 10.1111/j.1574-6941.2008.00587.x. Epub 2008 Sep 4. FEMS Microbiol Ecol. 2008. PMID: 18778275 - Complex Microbial Communities Drive Iron and Sulfur Cycling in Arctic Fjord Sediments.
Buongiorno J, Herbert LC, Wehrmann LM, Michaud AB, Laufer K, Røy H, Jørgensen BB, Szynkiewicz A, Faiia A, Yeager KM, Schindler K, Lloyd KG. Buongiorno J, et al. Appl Environ Microbiol. 2019 Jul 1;85(14):e00949-19. doi: 10.1128/AEM.00949-19. Print 2019 Jul 15. Appl Environ Microbiol. 2019. PMID: 31076435 Free PMC article. - Verrucomicrobia are candidates for polysaccharide-degrading bacterioplankton in an arctic fjord of Svalbard.
Cardman Z, Arnosti C, Durbin A, Ziervogel K, Cox C, Steen AD, Teske A. Cardman Z, et al. Appl Environ Microbiol. 2014 Jun;80(12):3749-56. doi: 10.1128/AEM.00899-14. Epub 2014 Apr 11. Appl Environ Microbiol. 2014. PMID: 24727271 Free PMC article. - Effects of shallow-water hydrothermal venting on biological communities of coastal marine ecosystems of the western Pacific.
Tarasov VG. Tarasov VG. Adv Mar Biol. 2006;50:267-421. doi: 10.1016/S0065-2881(05)50004-X. Adv Mar Biol. 2006. PMID: 16782453 Review. - Understanding Interaction Patterns within Deep-Sea Microbial Communities and Their Potential Applications.
Nawaz MZ, Subin Sasidharan R, Alghamdi HA, Dang H. Nawaz MZ, et al. Mar Drugs. 2022 Jan 28;20(2):108. doi: 10.3390/md20020108. Mar Drugs. 2022. PMID: 35200637 Free PMC article. Review.
Cited by
- Spatial variation and metabolic diversity of microbial communities in the surface sediments of the Mariana Trench.
Wang F, Zhang Y, Jing H, Liu H. Wang F, et al. Front Microbiol. 2022 Dec 5;13:1051999. doi: 10.3389/fmicb.2022.1051999. eCollection 2022. Front Microbiol. 2022. PMID: 36545198 Free PMC article. - Coupling Bacterioplankton Populations and Environment to Community Function in Coastal Temperate Waters.
Traving SJ, Bentzon-Tilia M, Knudsen-Leerbeck H, Mantikci M, Hansen JL, Stedmon CA, Sørensen H, Markager S, Riemann L. Traving SJ, et al. Front Microbiol. 2016 Sep 27;7:1533. doi: 10.3389/fmicb.2016.01533. eCollection 2016. Front Microbiol. 2016. PMID: 27729909 Free PMC article. - Polar front associated variation in prokaryotic community structure in Arctic shelf seafloor.
Nguyen TT, Landfald B. Nguyen TT, et al. Front Microbiol. 2015 Jan 23;6:17. doi: 10.3389/fmicb.2015.00017. eCollection 2015. Front Microbiol. 2015. PMID: 25667586 Free PMC article. - Metabolic Phenotyping of Marine Heterotrophs on Refactored Media Reveals Diverse Metabolic Adaptations and Lifestyle Strategies.
Forchielli E, Sher D, Segrè D. Forchielli E, et al. mSystems. 2022 Aug 30;7(4):e0007022. doi: 10.1128/msystems.00070-22. Epub 2022 Jul 20. mSystems. 2022. PMID: 35856685 Free PMC article. - Integrated metagenomic and metatranscriptomic analyses of microbial communities in the meso- and bathypelagic realm of north pacific ocean.
Wu J, Gao W, Johnson RH, Zhang W, Meldrum DR. Wu J, et al. Mar Drugs. 2013 Oct 11;11(10):3777-801. doi: 10.3390/md11103777. Mar Drugs. 2013. PMID: 24152557 Free PMC article.
References
- Abell, G. C. J., and J. P. Bowman. 2005. Colonization and community dynamics of class Flavobacteria on diatom detritus in experimental mesocosms based on Southern Ocean seawater. FEMS Microbiol. Ecol. 53:379-391. - PubMed
- Abell, G. C. J., and J. P. Bowman. 2005. Ecological and biogeographic relationships of class Flavobacteria in the Southern Ocean. FEMS Microbiol. Ecol. 51:265-277. - PubMed
- Alderkamp, A.-C., M. van Rijssel, and H. Bolhuis. 2007. Characterization of marine bacteria and the activity of their enzyme systems involved in degradation of the algal storage glucan laminarin. FEMS Microbiol. Ecol. 59:108-117. - PubMed
- Alonso, C., F. Warneke, R. Amann, and J. Pernthaler. 2007. High local and global diversity of Flavobacteria in marine plankton. Environ. Microbiol. 9:1253-1266. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases