Micropilot: automation of fluorescence microscopy-based imaging for systems biology - PubMed (original) (raw)
Micropilot: automation of fluorescence microscopy-based imaging for systems biology
Christian Conrad et al. Nat Methods. 2011 Mar.
Abstract
Quantitative microscopy relies on imaging of large cell numbers but is often hampered by time-consuming manual selection of specific cells. The 'Micropilot' software automatically detects cells of interest and launches complex imaging experiments including three-dimensional multicolor time-lapse or fluorescence recovery after photobleaching in live cells. In three independent experimental setups this allowed us to statistically analyze biological processes in detail and is thus a powerful tool for systems biology.
Figures
Figure 1
Schematic workflow of Micropilot. (a) After autofocussing different positions to find the best focal plane (yellow frame), low-resolution prescan images (optionally maximum _z_-dimension projections, gray frames) are presented to the automatic classification. If a cell is selected, a complex imaging protocol is executed; otherwise the system continues to prescan. After completion of the complex imaging protocol, the system loops back to prescan mode, continuing at the sample position where it stopped for the complex imaging mode. (b) Communication steps executed by the different microscope systems (red outlines) and the Micropilot software (blue outlines). The microscope sends the image path either via windows registry or socket interface to Micropilot. In the synchronous modes, each positive classification launches the complex imaging mode. In the asynchronous mode, microscope and Micropilot send and receive messages via transmission control protocol or internet protocol (TCP/IP), allowing classification of several different positions before launching the complex imaging protocol for a list of positions. (c) After reading the low-resolution image, Micropilot segments, extracts the feature set per object and classifies the cells during scanning to return eventually the positions of interest. After the criteria are met (time or number of positions) Micropilot deploys the complex imaging and the microscope switches back to prescan mode (a).
Figure 2
Assays of SEC31 and H2B-tubulin HeLa cells. (a) Examples for Hoechst-labeled (blue; DNA label) and SEC31-labeled (green) cells representing null or artifact and anaphase or telophase cells (insets, close-up images). Scale bars, 10 µm. (b) Confusion matrix of the prediction shows true positives (TP) horizontally against the predicted class vertically for cells. At edges the total numbers of the cells are given (overall total, 10,793 cells) corresponding to PPV = TP / (TP + false positives) and sensitivity = TP / (TP + false negatives). (c) Examples of null or artifact (left) and anaphase or telophase (right) cells stained with Hoechst (blue) and ERES spot (green) (50 slices of 0.2 µm). Scale bar, 10 µm. (d) Number of ERES spots of 91 anaphase cells to late-telophase cells plotted versus volume of nuclei, with exponential fit plotted. Red and blue data points correspond to the nuclei in the left and right images in c, respectively. (e) Example of negative control experiment (time resolution, 3 min; 30 slices of 1 µm; maximum projections) started after prophase recognition. Times indicated are after prophase recognition. Scale bar, 10 µm. (f) Spindle lengths after treatment with scrambled siRNA. (g) Example images after treatment with siRNA to CENPE, showing centrosome poles (arrows; left) for the first recognizable metaphase (acquisition as in e). Scale bar, 10 µm. (h) Normal mixture modeling of pole-pole distances in metaphase from 71 movies after treatment with siRNA to CENPE resulted in three distributions, which are shown as colored curves.
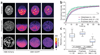
Figure 3
Examples and measurements of automatic FRAP on CBX1-EGFP cells. (a) After the automatic selection of an interphase or prophase cell with a trained prophase SVM classifier, a prebleached image was taken, followed by bleaching of the predefined upper half of the nucleus and subsequent time-lapse imaging with 2-s time resolution for 60 s (values in the lower images indicate time relative to bleaching). Scale bar, 5 µm. (b) Normalized intensities for CBX1-EGFP measured during fluorescence relaxation after photobleaching in interphase and prophase cells. We measured, normalized, averaged and plotted over time fluorescence intensities in bleached region of the nucleus. Error bars, s. (c) Recovery rates as box plots for interphase and prophase cell populations.
Similar articles
- [Parameters which affect the estimation of protein mobility by method FRAP in living cells on the example of protein fibrillarin].
Barygina VV, Mironova AA, Zatsepina OV. Barygina VV, et al. Tsitologiia. 2012;54(1):17-24. Tsitologiia. 2012. PMID: 22567896 Russian. - Analysis of Protein Kinetics Using Fluorescence Recovery After Photobleaching (FRAP).
Giakoumakis NN, Rapsomaniki MA, Lygerou Z. Giakoumakis NN, et al. Methods Mol Biol. 2017;1563:243-267. doi: 10.1007/978-1-4939-6810-7_16. Methods Mol Biol. 2017. PMID: 28324613 - Time-lapse, photoactivation, and photobleaching imaging of nucleolar assembly after mitosis.
Hernandez-Verdun D, Louvet E, Muro E. Hernandez-Verdun D, et al. Methods Mol Biol. 2013;1042:337-50. doi: 10.1007/978-1-62703-526-2_22. Methods Mol Biol. 2013. PMID: 23980017 - Visualization of TGN-endosome trafficking in mammalian and Drosophila cells.
Kametaka S, Waguri S. Kametaka S, et al. Methods Enzymol. 2012;504:255-71. doi: 10.1016/B978-0-12-391857-4.00013-6. Methods Enzymol. 2012. PMID: 22264539 Review. - Advances in fluorescent protein-based imaging for the analysis of plant endomembranes.
Held MA, Boulaflous A, Brandizzi F. Held MA, et al. Plant Physiol. 2008 Aug;147(4):1469-81. doi: 10.1104/pp.108.120147. Plant Physiol. 2008. PMID: 18678739 Free PMC article. Review. No abstract available.
Cited by
- Simplified equation to extract diffusion coefficients from confocal FRAP data.
Kang M, Day CA, Kenworthy AK, DiBenedetto E. Kang M, et al. Traffic. 2012 Dec;13(12):1589-600. doi: 10.1111/tra.12008. Epub 2012 Oct 10. Traffic. 2012. PMID: 22984916 Free PMC article. - Light microscopy applications in systems biology: opportunities and challenges.
Antony PM, Trefois C, Stojanovic A, Baumuratov AS, Kozak K. Antony PM, et al. Cell Commun Signal. 2013 Apr 11;11(1):24. doi: 10.1186/1478-811X-11-24. Cell Commun Signal. 2013. PMID: 23578051 Free PMC article. - Event-triggered STED imaging.
Alvelid J, Damenti M, Sgattoni C, Testa I. Alvelid J, et al. Nat Methods. 2022 Oct;19(10):1268-1275. doi: 10.1038/s41592-022-01588-y. Epub 2022 Sep 8. Nat Methods. 2022. PMID: 36076037 Free PMC article. - High-throughput micro-phenotyping measurements applied to assess stalk lodging in maize (Zea mays L.).
Zhang Y, Du J, Wang J, Ma L, Lu X, Pan X, Guo X, Zhao C. Zhang Y, et al. Biol Res. 2018 Oct 27;51(1):40. doi: 10.1186/s40659-018-0190-7. Biol Res. 2018. PMID: 30368254 Free PMC article. - Machine-Learning Optimization of Multiple Measurement Parameters Nonlinearly Affecting the Signal Quality.
Fujisaku T, So FTK, Igarashi R, Shirakawa M. Fujisaku T, et al. ACS Meas Sci Au. 2021 Jul 1;1(1):20-26. doi: 10.1021/acsmeasuresciau.1c00009. eCollection 2021 Aug 18. ACS Meas Sci Au. 2021. PMID: 36785732 Free PMC article.
References
- Pepperkok R, Ellenberg J. Nat. Rev. Mol. Cell Biol. 2006;7:690–696. - PubMed
- Ramo P, Sacher R, Snijder B, Begemann B, Pelkmans L. Bioinformatics. 2009;25:3028–3030. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases