De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks - PubMed (original) (raw)
De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks
Magdy M Mahfouz et al. Proc Natl Acad Sci U S A. 2011.
Abstract
Site-specific and rare cutting nucleases are valuable tools for genome engineering. The generation of double-strand DNA breaks (DSBs) promotes homologous recombination in eukaryotes and can facilitate gene targeting, additions, deletions, and inactivation. Zinc finger nucleases have been used to generate DSBs and subsequently, for genome editing but with low efficiency and reproducibility. The transcription activator-like family of type III effectors (TALEs) contains a central domain of tandem repeats that could be engineered to bind specific DNA targets. Here, we report the generation of a Hax3-based hybrid TALE nuclease with a user-selected DNA binding specificity. We show that the engineered TALE nuclease can bind to its target sequence in vitro and that the homodimeric TALE nuclease can cleave double-stranded DNA in vitro if the DNA binding sites have the proper spacing and orientation. Transient expression assays in tobacco leaves suggest that the hybrid nuclease creates DSB in its target sequence, which is subsequently repaired by nonhomologous end-joining repair. Taken together, our data show the feasibility of engineering TALE-based hybrid nucleases capable of generating site-specific DSBs and the great potential for site-specific genome modification in plants and eukaryotes in general.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
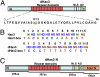
Structural representation of Hax3 and dHax3.N proteins and their DNA target specificity. (A) Hax3 TALE contains a central repeat domain shown in red, a nuclear localization signal (NLS) shown in yellow, and a transcriptional activation domain (AD) shown in green. The central repeat domain consists of 11.5 repeat units, and each repeat unit is composed of 34 aa; a representative first repeat sequence is shown below, and the RVD is shown in blue. (B) RVDs of the 11.5 repeat units of Hax3 and dHax3 with their DNA target sequences shown below. (C) Structural representation of the dHax3.N showing the fusion of 196 aa of the FokI cleavage domain at the C terminus of dHax3.
Fig. 2.
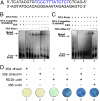
Binding of dHax3 and dHax3.N to their target sequence and in vivo transcriptional activation. (A) Probe containing the EBE sequence (shown in blue) used in the EMSA. (B) EMSA showing dHax3 binding to its target sequence. Panel 1 shows that the thioredoxin tag does not bind to the biotin-labeled probe. Panel 2 shows that the dHax3 protein binds to the biotin-labeled probe sequence in a concentration-dependent manner. Panel 3 indicates that the unlabeled probe competitively reduces the binding of the biotin-labeled probe. (C) EMSA showing the binding of dHax3.N to its target DNA; panel 2 shows that dHax3.N binds to its target in a concentration-dependent manner, and panel 3 shows competition of binding by the unlabeled probe. (D) dHax3 and dHax3.N activate the RD29A promoter. Panels 1 and 2 serve as controls and indicate no uidA activity. Panel 3 indicates the background activity of RD29A promoter. Panels 4 and 5 show transcriptional activation of RD29A promoter by dHax3 and dHax3.N, respectively. Panel 6 shows a positive control of 35S::uidA. The experiment was repeated three times with similar results; 60 plants were used for each experiment (10 plants/panel).
Fig. 3.
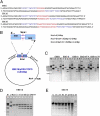
In vitro digestion of DNA target with spacers of different lengths. (A) DNA target sequence with 6-, 10-, and 16-bp spacer EBE sequences are shown in blue, and the spacer sequences are shown in red. (B) Representation of the circular pCRII-TOPO plasmid containing two EBE elements in a tail to tail orientation and the predicted cleavage sites for SmaI and NcoI enzymes. (C) In vitro digestion of DNA targets with different spacer lengths. In all EBE panels, lane 1 is undigested plasmid. Lane 2 is NcoI-linearized plasmid. Lane 3 is digested with NcoI and SmaI. Lane 4 is 500 ng NcoI-linearized plasmid digested with 50 ng dHax3.N for 30 min at 37 °C. Lane 4 in panel EBE6 does not have the predicted digestion products, indicating no cleavage activity by dHax3.N with a 6-bp spacer between the two EBEs. Lane 4 in the EBE10 panel shows minimal activity of the enzyme, resulting in a very small amount of the predicted products. Lane 4 of panel EBE16 shows substantial activity of dHax3.N, indicating an optimal spacer length. In lane 4 of panel EBE16 T/A, the replacement of T by A in the last nucleotide in the target sequence results in minimal or no activity of dHax3.N. In lane 4 of EBE16ΔT0, the removal of T in the target sequence that corresponds to repeat 0 leads to minimal or no dHax3.N activity. M, 1-kb marker. (D) Kinetics of cleavage of EBE16 by 50 ng dHax3.N. U, undigested plasmid; NcoI, NcoI-linearized DNA. (E) Effect of temperature on dHax3.N activity.
Fig. 4.
dHax3.N activity in vivo. (A) The sequence of the EBE target is preceded by the initiation codon (ATG) and contains a TGA stop codon in the spacer region. The EBE target sequence is fused in frame to the uidA coding sequence. (B) dHax3.N activity in tobacco transient assays. Panels 1 and 2 show no activity of 35S::EBE.TGA.spacer.EBE.uidA or 35S::dHax3, respectively. Panel 3 shows uidA activity when 35S::EBE.TGA.spacer.EBE.uidA was coinfiltrated with 35S::dHax3.N. Panel 4 is a positive control that shows the activity of 35S::uidA. The experiment was repeated three times with similar results; 40 plants were used for each experiment (10 plants/panel).
Fig. 5.
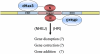
Diagram of the dHax3.N hybrid nuclease with its DNA target and potential applications. dHax3 TALE provides a DNA binding module, and the FokI cleavage domain provides the nuclease activity. Two monomeric subunits of dHax3.N are required for the cleavage activity. DSBs created by dHax3.N could be used to achieve gene disruption, correction, and addition mediated by NHEJ or HR pathways.
Similar articles
- Off-target effects of engineered nucleases.
Yee JK. Yee JK. FEBS J. 2016 Sep;283(17):3239-48. doi: 10.1111/febs.13760. Epub 2016 Jun 6. FEBS J. 2016. PMID: 27208701 Review. - Rapid and highly efficient construction of TALE-based transcriptional regulators and nucleases for genome modification.
Li L, Piatek MJ, Atef A, Piatek A, Wibowo A, Fang X, Sabir JS, Zhu JK, Mahfouz MM. Li L, et al. Plant Mol Biol. 2012 Mar;78(4-5):407-16. doi: 10.1007/s11103-012-9875-4. Epub 2012 Jan 22. Plant Mol Biol. 2012. PMID: 22271303 Free PMC article. - Targeting DNA double-strand breaks with TAL effector nucleases.
Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Christian M, et al. Genetics. 2010 Oct;186(2):757-61. doi: 10.1534/genetics.110.120717. Epub 2010 Jul 26. Genetics. 2010. PMID: 20660643 Free PMC article. - Site-specific gene targeting using transcription activator-like effector (TALE)-based nuclease in Brassica oleracea.
Sun Z, Li N, Huang G, Xu J, Pan Y, Wang Z, Tang Q, Song M, Wang X. Sun Z, et al. J Integr Plant Biol. 2013 Nov;55(11):1092-103. doi: 10.1111/jipb.12091. Epub 2013 Sep 18. J Integr Plant Biol. 2013. PMID: 23870552 - Designed nucleases for targeted genome editing.
Lee J, Chung JH, Kim HM, Kim DW, Kim H. Lee J, et al. Plant Biotechnol J. 2016 Feb;14(2):448-62. doi: 10.1111/pbi.12465. Epub 2015 Sep 15. Plant Biotechnol J. 2016. PMID: 26369767 Review.
Cited by
- Genome editing in Sub-Saharan Africa: a game-changing strategy for climate change mitigation and sustainable agriculture.
Amoah P, Oumarou Mahamane AR, Byiringiro MH, Mahula NJ, Manneh N, Oluwasegun YR, Assfaw AT, Mukiti HM, Garba AD, Chiemeke FK, Bernard Ojuederie O, Olasanmi B. Amoah P, et al. GM Crops Food. 2024 Dec 31;15(1):279-302. doi: 10.1080/21645698.2024.2411767. Epub 2024 Oct 31. GM Crops Food. 2024. PMID: 39481911 Free PMC article. Review. - Unconstrained Precision Mitochondrial Genome Editing with αDdCBEs.
Castillo SR, Simone BW, Clark KJ, Devaux P, Ekker SC. Castillo SR, et al. Hum Gene Ther. 2024 Oct;35(19-20):798-813. doi: 10.1089/hum.2024.073. Epub 2024 Sep 24. Hum Gene Ther. 2024. PMID: 39212664 - Crop bioengineering via gene editing: reshaping the future of agriculture.
Atia M, Jiang W, Sedeek K, Butt H, Mahfouz M. Atia M, et al. Plant Cell Rep. 2024 Mar 18;43(4):98. doi: 10.1007/s00299-024-03183-1. Plant Cell Rep. 2024. PMID: 38494539 Free PMC article. Review. - Outlook on genome editing application to cattle.
Gim GM, Jang G. Gim GM, et al. J Vet Sci. 2024 Jan;25(1):e10. doi: 10.4142/jvs.23133. J Vet Sci. 2024. PMID: 38311323 Free PMC article. Review. - CRISPR technology towards genome editing of the perennial and semi-perennial crops citrus, coffee and sugarcane.
Prado GS, Rocha DC, Dos Santos LN, Contiliani DF, Nobile PM, Martinati-Schenk JC, Padilha L, Maluf MP, Lubini G, Pereira TC, Monteiro-Vitorello CB, Creste S, Boscariol-Camargo RL, Takita MA, Cristofani-Yaly M, de Souza AA. Prado GS, et al. Front Plant Sci. 2024 Jan 8;14:1331258. doi: 10.3389/fpls.2023.1331258. eCollection 2023. Front Plant Sci. 2024. PMID: 38259920 Free PMC article. Review.
References
- Pennisi E. Sowing the seeds for the ideal crop. Science. 2010;327:802–803. - PubMed
- Weinthal D, Tovkach A, Zeevi V, Tzfira T. Genome editing in plant cells by zinc finger nucleases. Trends Plant Sci. 2010;15:308–321. - PubMed
- Cathomen T, Joung JK. Zinc-finger nucleases: The next generation emerges. Mol Ther. 2008;16:1200–1207. - PubMed
- Kim YG, Shi Y, Berg JM, Chandrasegaran S. Site-specific cleavage of DNA-RNA hybrids by zinc finger/FokI cleavage domain fusions. Gene. 1997;203:43–49. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources