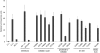Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3' to 5' exonuclease activity essential for immune suppression - PubMed (original) (raw)
Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3' to 5' exonuclease activity essential for immune suppression
Kathryn M Hastie et al. Proc Natl Acad Sci U S A. 2011.
Abstract
Lassa fever virus, a member of the family Arenaviridae, is a highly endemic category A pathogen that causes 300,000-500,000 infections per year in Western Africa. The arenaviral nucleoprotein NP has been implicated in suppression of the host innate immune system, but the mechanism by which this occurs has remained elusive. Here we present the crystal structure at 1.5 Å of the immunosuppressive C-terminal portion of Lassa virus NP and illustrate that, unexpectedly, its 3D fold closely mimics that of the DEDDh family of exonucleases. Accompanying biochemical experiments illustrate that NP indeed has a previously unknown, bona fide exonuclease activity, with strict specificity for double-stranded RNA substrates. We further demonstrate that this exonuclease activity is essential for the ability of NP to suppress translocation of IFN regulatory factor 3 and block activation of the innate immune system. Thus, the nucleoprotein is a viral exonuclease with anti-immune activity, and this work provides a unique opportunity to combat arenaviral infections.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
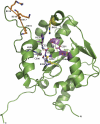
Fig. 1.
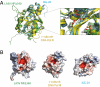
Structure of the C-terminal, immunosuppressive domain of LASV NP. (A) Cartoon representation of LASV NPΔ340. The basic arm includes residues Lys516, Lys517, Lys518, and Arg519. (B) A single zinc is coordinated by Glu399, His506, Cys509, and Cys529.
Fig. 2.
Superimposition of LASV NP and known DEDD exonucleases. (A) Structural comparison of NPΔ340 and two known DEDDh exonucleases. NPΔ340 is colored green, ISG-20 (PDB ID 1WLJ; ref. 27) is colored cyan, and the E. coli DNA pol IIIε (PDB ID 2GUI; ref. 28) is colored yellow. Inset shows a close-up view of the superimposed DEDDh residues of the active site. Numbered residues reflect those of LASV NP. (B) Electrostatic surface potential calculated with APBS (the Adaptive Poisson-Boltzmann Solver) software (49) shows that each exonuclease has an acidic active site and highlights the basic arm of LASV NP. Positive surface is colored blue; negative surface is colored red with limits ± 10 kT/e.
Fig. 3.
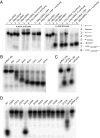
Ribonucleolytic activity of LASV NP. (A) Substrate specificity. NPΔ340 was incubated with different 32P-labeled substrates, and the reaction products were analyzed by denaturing PAGE and autoradiography. (Left) Migration patterns of each nucleic acid substrate alone (in the absence of NPΔ340 exonuclease). (Right) Migration of each substrate when incubated with NPΔ340 for 15 min. A schematic for each substrate is also shown, with the location of the 32P label indicated by an asterisk. Sequences for each substrate can be found in
Table S2
. (B) Time course of exonuclease activity. The 18-bp blunt-ended dsRNA is increasingly digested by wild-type NPΔ340 from 0 to 15 min. (C) Comparison of ribonucleolytic activity of the N terminus of NP (NP1–340) and full-length NP (NP-FL). The 18-bp blunt-ended dsRNA is digested by full-length NP and NPΔ340, whereas NP1–340 does not have exonuclease activity. (D) Effects of mutations to active site and proximal residues on ribonucleolytic activity. Wild-type and NPΔ340 point mutants were incubated with 18-bp blunt-ended dsRNA for 15 min, and products were analyzed by PAGE. QuadA designates a quadruple alanine mutation in residues K516K517K518R519. Note that all point mutations to residues in or near the exonuclease active site and the Zn coordination site, save R393, abrogate exonuclease activity, leaving the dsRNA undigested. The QuadA mutation to the basic arm partially diminishes exonuclease activity.
Fig. 4.
Cartoon representation of NPΔ340 and location of residues important for the exonuclease and anti-IFN activities. Residues corresponding to the DEDDh motif are colored magenta, two Arg residues proximal to the active site are colored yellow, residues involved in coordination of the Zn2+ atom are colored blue, and residues corresponding to the basic arm are colored orange.
Fig. 5.
Sendai virus-mediated activation of an IRF-3 promoter. Full-length NP (NP-FL), the N-terminal domain of NP (NP1–340), the C-terminal domain of NP (NPΔ340), and amino acid substitutions of full-length NP were assayed for inhibition of Sendai virus-mediated activation of an IRF-3–dependent promoter. Values displayed reflect relative luminescence units corrected to an uninfected, empty vector control. E(−) and E(+) indicate an uninfected empty vector control and an infected empty vector control, respectively. Note that the wild-type function of NP is to block IRF-3 translocation and therefore block reporter luminescence.
Similar articles
- Structures of arenaviral nucleoproteins with triphosphate dsRNA reveal a unique mechanism of immune suppression.
Jiang X, Huang Q, Wang W, Dong H, Ly H, Liang Y, Dong C. Jiang X, et al. J Biol Chem. 2013 Jun 7;288(23):16949-16959. doi: 10.1074/jbc.M112.420521. Epub 2013 Apr 24. J Biol Chem. 2013. PMID: 23615902 Free PMC article. - Cap binding and immune evasion revealed by Lassa nucleoprotein structure.
Qi X, Lan S, Wang W, Schelde LM, Dong H, Wallat GD, Ly H, Liang Y, Dong C. Qi X, et al. Nature. 2010 Dec 9;468(7325):779-83. doi: 10.1038/nature09605. Epub 2010 Nov 17. Nature. 2010. PMID: 21085117 Free PMC article. - Lassa Virus, but Not Highly Pathogenic New World Arenaviruses, Restricts Immunostimulatory Double-Stranded RNA Accumulation during Infection.
Mateer EJ, Maruyama J, Card GE, Paessler S, Huang C. Mateer EJ, et al. J Virol. 2020 Apr 16;94(9):e02006-19. doi: 10.1128/JVI.02006-19. Print 2020 Apr 16. J Virol. 2020. PMID: 32051278 Free PMC article. - Hiding the evidence: two strategies for innate immune evasion by hemorrhagic fever viruses.
Hastie KM, Bale S, Kimberlin CR, Saphire EO. Hastie KM, et al. Curr Opin Virol. 2012 Apr;2(2):151-6. doi: 10.1016/j.coviro.2012.01.003. Epub 2012 Jan 28. Curr Opin Virol. 2012. PMID: 22482712 Free PMC article. Review. - Innate immune response to arenaviral infection: a focus on the highly pathogenic New World hemorrhagic arenaviruses.
Koma T, Huang C, Kolokoltsova OA, Brasier AR, Paessler S. Koma T, et al. J Mol Biol. 2013 Dec 13;425(24):4893-903. doi: 10.1016/j.jmb.2013.09.028. Epub 2013 Sep 26. J Mol Biol. 2013. PMID: 24075870 Free PMC article. Review.
Cited by
- A recently isolated Lassa virus from Mali demonstrates atypical clinical disease manifestations and decreased virulence in cynomolgus macaques.
Safronetz D, Strong JE, Feldmann F, Haddock E, Sogoba N, Brining D, Geisbert TW, Scott DP, Feldmann H. Safronetz D, et al. J Infect Dis. 2013 Apr 15;207(8):1316-27. doi: 10.1093/infdis/jit004. Epub 2013 Jan 9. J Infect Dis. 2013. PMID: 23303805 Free PMC article. - Self-association of lymphocytic choriomeningitis virus nucleoprotein is mediated by its N-terminal region and is not required for its anti-interferon function.
Ortiz-Riaño E, Cheng BY, de la Torre JC, Martínez-Sobrido L. Ortiz-Riaño E, et al. J Virol. 2012 Mar;86(6):3307-17. doi: 10.1128/JVI.05503-11. Epub 2012 Jan 18. J Virol. 2012. PMID: 22258244 Free PMC article. - Monomeric nucleoprotein of influenza A virus.
Chenavas S, Estrozi LF, Slama-Schwok A, Delmas B, Di Primo C, Baudin F, Li X, Crépin T, Ruigrok RW. Chenavas S, et al. PLoS Pathog. 2013 Mar;9(3):e1003275. doi: 10.1371/journal.ppat.1003275. Epub 2013 Mar 28. PLoS Pathog. 2013. PMID: 23555270 Free PMC article. - Host Cell Restriction Factors of Bunyaviruses and Viral Countermeasures.
Lerolle S, Freitas N, Cosset FL, Legros V. Lerolle S, et al. Viruses. 2021 Apr 28;13(5):784. doi: 10.3390/v13050784. Viruses. 2021. PMID: 33925004 Free PMC article. Review. - Genome encapsidation by orthobunyavirus nucleoproteins.
Zheng W, Tao YJ. Zheng W, et al. Proc Natl Acad Sci U S A. 2013 May 28;110(22):8769-70. doi: 10.1073/pnas.1306838110. Epub 2013 May 21. Proc Natl Acad Sci U S A. 2013. PMID: 23696659 Free PMC article. No abstract available.
References
- Buchmeier MJ, de la Torre JC, Peters CJ. Arenaviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th Ed. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1791–1827.
- Haas WH, et al. Imported Lassa fever in Germany: Surveillance and management of contact persons. Clin Infect Dis. 2003;36:1254–1258. - PubMed
- Holmes GP, et al. Lassa fever in the United States. Investigation of a case and new guidelines for management. N Engl J Med. 1990;323:1120–1123. - PubMed
- Jamieson DJ, Kourtis AP, Bell M, Rasmussen SA. Lymphocytic choriomeningitis virus: An emerging obstetric pathogen? Am J Obstet Gynecol. 2006;194:1532–1536. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous