Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression - PubMed (original) (raw)
Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression
Christian Boehler et al. Proc Natl Acad Sci U S A. 2011.
Abstract
The ADP ribosyl transferase [poly(ADP-ribose) polymerase] ARTD3(PARP3) is a newly characterized member of the ARTD(PARP) family that catalyzes the reaction of ADP ribosylation, a key posttranslational modification of proteins involved in different signaling pathways from DNA damage to energy metabolism and organismal memory. This enzyme shares high structural similarities with the DNA repair enzymes PARP1 and PARP2 and accordingly has been found to catalyse poly(ADP ribose) synthesis. However, relatively little is known about its in vivo cellular properties. By combining biochemical studies with the generation and characterization of loss-of-function human and mouse models, we describe PARP3 as a newcomer in genome integrity and mitotic progression. We report a particular role of PARP3 in cellular response to double-strand breaks, most likely in concert with PARP1. We identify PARP3 as a critical player in the stabilization of the mitotic spindle and in telomere integrity notably by associating and regulating the mitotic components NuMA and tankyrase 1. Both functions open stimulating prospects for specifically targeting PARP3 in cancer therapy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
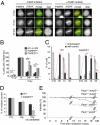
PARP3 is involved in DNA damage response. (A) Endogenous PARP3 accumulates at laser-induced DNA damage sites detected by an anti-γH2AX antibody. Wide-field fluorescence images of Cos1 either untreated or treated with the PARP inhibitor. Fixation and immunostaining was performed at indicated time points after laser microirradiation. (Scale bar, 5 μm.) (B) Spontaneous accumulation and persistence of X-ray-induced γH2AX foci in PARP3kd cells. Quantification of the percentage of cells displaying γH2AX foci. An average of 500 cells per cell line were scored in >20 randomly selected fields. (C) Histogram showing the percentage of unrepaired (with COMET) vs. repaired (round shaped) control (ctl1) and PARP3kd1 cells as a function of time after X-irradiation. An average of 50 cells was scored for each time point. For B and C, results are averages from three independent experiments. Mean values ± SD are indicated. *P < 0.05, **P < 0.01, ***P < 0.001. (D) Clonogenic survival of untreated (CTL) or 1 Gy-X-irradiated control (ctl) and PARP3kd1 cells in the absence or in the presence of Ku-0058948 (100 nM). Experiments were performed three times, giving similar results. Mean values of triplicates ± SD are indicated. *P < 0.05. (E) Increased radiosensitivity in Parp1−/−;Parp3−/− double-knockout mice. Kaplan–Meier survival curve after 4-Gy whole-body X-irradiation. Parp1+/+;Parp3+/+ (n = 9), Parp1+/+;Parp3−/− (n = 9), Parp1−/−;Parp3+/+ (n = 9), Parp1−/−;Parp3−/− (n = 11).
Fig. 2.
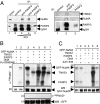
Physical and functional association of PARP3 with NuMA and tankyrase 1. (A Left) Coimmunoprecipitation of NuMA with PARP3 after increasing stringency conditions of washing buffers. Cos1 cell extracts were immunoprecipitated with a control antibody (lane 2) or an anti-PARP3 antibody (lanes 3 and 4) and analyzed by Western blotting using successively anti-NuMA and anti-PARP3 antibodies. Input corresponds to 1/13 of the total amount of cell extract used for immunoprecipitation. Lane 5, purified recombinant PARP3 (10 ng). (A Right) Coimmunoprecipitation of tankyrase 1 and NuMA with PARP3. Cos1 cell extracts were immunoprecipitated with a control antibody (lane 1) or an anti-PARP3 antibody (lane 2) and analyzed by Western blotting using successively anti-NuMA, anti-tankyrase 1 and anti-PARP3 antibodies. Lane 3, purified recombinant PARP3 (10 ng). (B) PARP3 induces the ADP ribosylation of NuMA both directly and through tankyrase 1. (a and b) Immunopurified GFP-NuMA was incubated with purified PARP3 and/or tankyrase 1 (TNKS1) as indicated in PARP activity buffer. The addition of Ku-0058948 (250 nM) significantly inhibits PARP3 but not tankyrase 1 (lane 7 vs. lanes 2 and 3). (c and d) In similar experimental conditions as above, no ADP ribosylation of GFP alone was detected. (a and c) Autoradiography. (b and d) Immunopurified GFP or GFP-NuMA were analyzed by Western blotting using an anti-GFP antibody. (C) PARP3 stimulates the auto-ADP ribosylation of tankyrase 1. Immunopurified GFP-NuMA were incubated with purified PARP3 or tankyrase 1 and assayed for PARP activity as above. The addition of XAV-939 (500 nM) inhibits efficiently tankyrase 1 but not PARP3 (compare lanes 4 with 3, and 2 with 1). (a) Autoradiography. (b) Immunopurified GFP-NuMA was analyzed by Western blotting using an anti-GFP antibody.
Fig. 3.
PARP3 is required for efficient mitotic progression. (A) Histogram showing an increase in the percentage of PARP3kd1-GFP-H2B mitotic cells displaying a delay in prometaphase-to-metaphase transition or a delay in metaphase compared with normal mitotic progression in ctl1-GFP-H2B. For each step, >100 mitoses were scored by life cell microscopy. ***P < 0.0001. (B) Representative time-lapse video live-cell imaging of ctl1-GFP-H2B and PARP3kd-GFP-H2B cells. Whereas a normal mitotic progression is observed in the control cell, PARP3kd-GFP-H2B remain delayed for up to 80 min in the prometaphase-to-metaphase transition. (C) Representative time-lapse video live-cell imaging of the PARP3kd-GFP-H2B cell line showing either a delay of ∼220 min in metaphase (Left) or a metaphase arrest resulting in mitotic cell death (Right). (D) Frequencies of mitotic cell death in ctl1-GFP-H2B and PARP3kd-GFP-H2B cells. More than 100 mitoses were scored by life cell microscopy. ***P < 0.0001.
Fig. 4.
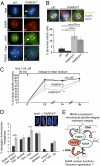
PARP3 is required for microtubule spindle dynamics and telomere integrity. (A) Spindle defects in PARP3-depleted cells. (a–f) Representative images of NuMA (red)/α tubulin (green) coimmunostaining of control (ctl1) and PARP3kd metaphases, counterstained with DAPI (blue). Note the splayed microtubules in PARP3kd cells (arrow) compared with a focused spindle in ctl1 cells, despite a wild-type–like accumulation of NuMA to the spindle poles (e and f vs. b and c). (g and h) Representative images of tankyrase 1 (TNKS1) (green) immunostaining of control (ctl1) and PARP3kd metaphases showing a normal targeting of TNKS1 to the spindle poles. DNA is counterstained with DAPI (blue). (B) Accumulation of abnormal mitotic cells in PARP3kd cells. Percentage of abnormal mitotic figures with supernumerary spindle poles in PARP3kd cells (b and c) vs. ctl cells (a) determined by coimmunodetection of α-tubulin and NuMA and DNA staining with DAPI. An average of 65 mitotic cells were scored per cell line in >20 randomly selected immunofluorescence fields. Results are averages of five independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001. (C) Spindle microtubule regrowth is delayed in PARP3kd cells. Microtubules were depolymerized by nocodazole treatment as indicated, and repolymerized at 37 °C. At the indicated time points, cells were fixed, coimmunostained for NuMA and α-tubulin, and scored for the formation of regular compact spindle. Over 45 cells were scored for each independent cell line. Results are averages from three independent experiments. *P < 0.05, **P < 0.01. (D) Spontaneous increase in sister telomere fusions and sister telomere loss in PARP3kd1 compared with ctl1 cells. Telomere aberrations were detected by FISH on metaphase spreads and expressed as percentages of damaged chromosomes per metaphase. (Insets) Sister telomere fusions (b and c) and telomere loss (d) identified in PARP3kd metaphases compared with normal telomeres (a) observed in ctl1 cells. (E) Working model posing the dual functions of PARP3 in association with NuMA. Within the mitotic protein network containing tankyrase 1 and NuMA, PARP3 stimulates (indicated by +++) the tankyrase 1 catalyzed auto-ADP ribosylation and hetero-ADP ribosylation of NuMA to favor telomere integrity and spindle dynamics in a DNA-independent manner. In addition, PARP3 is able to directly ADP ribosylate NuMA in a DNA-dependent manner, a possible means of regulating its functions in the interphase nucleus. Gray arrow, tankyrase 1-catalyzed poly(ADP ribosyl)ation; red arrow, PARP3-catalyzed ADP ribosylation.
Comment in
- PARP-3, a DNA-dependent PARP with emerging roles in double-strand break repair and mitotic progression.
Boehler C, Dantzer F. Boehler C, et al. Cell Cycle. 2011 Apr 1;10(7):1023-4. doi: 10.4161/cc.10.7.15169. Epub 2011 Apr 1. Cell Cycle. 2011. PMID: 21358266 No abstract available.
Similar articles
- Characterization of DNA ADP-ribosyltransferase activities of PARP2 and PARP3: new insights into DNA ADP-ribosylation.
Zarkovic G, Belousova EA, Talhaoui I, Saint-Pierre C, Kutuzov MM, Matkarimov BT, Biard D, Gasparutto D, Lavrik OI, Ishchenko AA. Zarkovic G, et al. Nucleic Acids Res. 2018 Mar 16;46(5):2417-2431. doi: 10.1093/nar/gkx1318. Nucleic Acids Res. 2018. PMID: 29361132 Free PMC article. - Mitotic functions of poly(ADP-ribose) polymerases.
Slade D. Slade D. Biochem Pharmacol. 2019 Sep;167:33-43. doi: 10.1016/j.bcp.2019.03.028. Epub 2019 Mar 22. Biochem Pharmacol. 2019. PMID: 30910692 Free PMC article. Review. - PARP3 inhibitors ME0328 and olaparib potentiate vinorelbine sensitization in breast cancer cell lines.
Sharif-Askari B, Amrein L, Aloyz R, Panasci L. Sharif-Askari B, et al. Breast Cancer Res Treat. 2018 Nov;172(1):23-32. doi: 10.1007/s10549-018-4888-6. Epub 2018 Jul 23. Breast Cancer Res Treat. 2018. PMID: 30039287 - NuMA is a major acceptor of poly(ADP-ribosyl)ation by tankyrase 1 in mitosis.
Chang W, Dynek JN, Smith S. Chang W, et al. Biochem J. 2005 Oct 15;391(Pt 2):177-84. doi: 10.1042/BJ20050885. Biochem J. 2005. PMID: 16076287 Free PMC article. - Poly(ADP-ribose) polymerases in double-strand break repair: focus on PARP1, PARP2 and PARP3.
Beck C, Robert I, Reina-San-Martin B, Schreiber V, Dantzer F. Beck C, et al. Exp Cell Res. 2014 Nov 15;329(1):18-25. doi: 10.1016/j.yexcr.2014.07.003. Epub 2014 Jul 10. Exp Cell Res. 2014. PMID: 25017100 Review.
Cited by
- Kdm4b histone demethylase is a DNA damage response protein and confers a survival advantage following γ-irradiation.
Young LC, McDonald DW, Hendzel MJ. Young LC, et al. J Biol Chem. 2013 Jul 19;288(29):21376-21388. doi: 10.1074/jbc.M113.491514. Epub 2013 Jun 6. J Biol Chem. 2013. PMID: 23744078 Free PMC article. - Exploring molecular pathways of triple-negative breast cancer.
Ossovskaya V, Wang Y, Budoff A, Xu Q, Lituev A, Potapova O, Vansant G, Monforte J, Daraselia N. Ossovskaya V, et al. Genes Cancer. 2011 Sep;2(9):870-9. doi: 10.1177/1947601911432496. Genes Cancer. 2011. PMID: 22593799 Free PMC article. - Identification of differentially expressed genes associated with the enhancement of X-ray susceptibility by RITA in a hypopharyngeal squamous cell carcinoma cell line (FaDu).
Luan J, Li X, Guo R, Liu S, Luo H, You Q. Luan J, et al. Radiol Oncol. 2016 Feb 22;50(2):168-74. doi: 10.1515/raon-2016-0010. eCollection 2016 Jun 1. Radiol Oncol. 2016. PMID: 27247549 Free PMC article. - Poly (ADP-ribose) polymerase inhibitors: recent advances and future development.
Scott CL, Swisher EM, Kaufmann SH. Scott CL, et al. J Clin Oncol. 2015 Apr 20;33(12):1397-406. doi: 10.1200/JCO.2014.58.8848. Epub 2015 Mar 16. J Clin Oncol. 2015. PMID: 25779564 Free PMC article. Review. - Crosstalk between poly(ADP-ribose) polymerase and sirtuin enzymes.
Cantó C, Sauve AA, Bai P. Cantó C, et al. Mol Aspects Med. 2013 Dec;34(6):1168-201. doi: 10.1016/j.mam.2013.01.004. Epub 2013 Jan 25. Mol Aspects Med. 2013. PMID: 23357756 Free PMC article. Review.
References
- Schreiber V, Dantzer F, Ame J-C, de Murcia G. Poly(ADP-ribose): Novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. - PubMed
- Hottiger MO, Hassa PO, Lüscher B, Schüler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35:208–219. - PubMed
- Johansson M. A human poly(ADP-ribose) polymerase gene family (ADPRTL): cDNA cloning of two novel poly(ADP-ribose) polymerase homologues. Genomics. 1999;57:442–445. - PubMed
- Urbánek P, Paces J, Králová J, Dvorák M, Paces V. Cloning and expression of PARP-3 (Adprt3) and U3-55k, two genes closely linked on mouse chromosome 9. Folia Biol (Praha) 2002;48:182–191. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous