An accurate sequentially Markov conditional sampling distribution for the coalescent with recombination - PubMed (original) (raw)
An accurate sequentially Markov conditional sampling distribution for the coalescent with recombination
Joshua S Paul et al. Genetics. 2011 Apr.
Abstract
The sequentially Markov coalescent is a simplified genealogical process that aims to capture the essential features of the full coalescent model with recombination, while being scalable in the number of loci. In this article, the sequentially Markov framework is applied to the conditional sampling distribution (CSD), which is at the core of many statistical tools for population genetic analyses. Briefly, the CSD describes the probability that an additionally sampled DNA sequence is of a certain type, given that a collection of sequences has already been observed. A hidden Markov model (HMM) formulation of the sequentially Markov CSD is developed here, yielding an algorithm with time complexity linear in both the number of loci and the number of haplotypes. This work provides a highly accurate, practical approximation to a recently introduced CSD derived from the diffusion process associated with the coalescent with recombination. It is empirically demonstrated that the improvement in accuracy of the new CSD over previously proposed HMM-based CSDs increases substantially with the number of loci. The framework presented here can be adopted in a wide range of applications in population genetics, including imputing missing sequence data, estimating recombination rates, and inferring human colonization history.
Figures
Figure 1.—
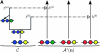
Illustration of the corresponding genealogical and sequential interpretations for a realization of . The three loci of each haplotype are each represented by a solid circle, with the color indicating the allelic type at that locus. The trunk genealogy 𝒜*(n) and conditional genealogy C are indicated. Time is represented vertically, with the present (time 0) at the bottom of the illustration. (A) The genealogical interpretation: Mutation events, along with the locus and resulting haplotype, are indicated by small arrows. Recombination events, and the resulting haplotype, are indicated by branching events in C. Absorption events, and the corresponding absorption time [t(a) and t(b)] and haplotype [h(a) and h(b), respectively], are indicated by dotted-dashed horizontal lines. (B) The corresponding sequential interpretation: The marginal genealogies at the first, second, and third locus (_S_1, _S_2, and _S_3) are emphasized as dotted, dashed, and solid lines, respectively. Mutation events at each locus, along with resulting allele, are indicated by small arrows. Absorption events at each locus are indicated by horizontal lines.
Figure 1.—
Illustration of the corresponding genealogical and sequential interpretations for a realization of . The three loci of each haplotype are each represented by a solid circle, with the color indicating the allelic type at that locus. The trunk genealogy 𝒜*(n) and conditional genealogy C are indicated. Time is represented vertically, with the present (time 0) at the bottom of the illustration. (A) The genealogical interpretation: Mutation events, along with the locus and resulting haplotype, are indicated by small arrows. Recombination events, and the resulting haplotype, are indicated by branching events in C. Absorption events, and the corresponding absorption time [t(a) and t(b)] and haplotype [h(a) and h(b), respectively], are indicated by dotted-dashed horizontal lines. (B) The corresponding sequential interpretation: The marginal genealogies at the first, second, and third locus (_S_1, _S_2, and _S_3) are emphasized as dotted, dashed, and solid lines, respectively. Mutation events at each locus, along with resulting allele, are indicated by small arrows. Absorption events at each locus are indicated by horizontal lines.
Figure 2.—
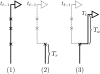
Illustration of the (Markov) process for sampling the absorption time _T_ℓ given the absorption time _T_ℓ−1 = t_ℓ−1. In step 1, recombination breakpoints are realized as a Poisson process with rate ρ_b/2 on the marginal conditional genealogy with absorption time _t_ℓ−1. In step 2, the lineage branching from each breakpoint associated with locus ℓ−1 is removed, so that only the lineage more recent than the first breakpoint, at time _T_r, remains. In step 3, the lineage branching from the first recombination breakpoint associated with locus ℓ is absorbed after time _T_a distributed exponentially with rate n/2. Thus, _T_ℓ = _T_r + _T_a.
Figure 3.—
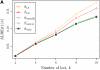
Absolute log-ratio error (ALRErr) of various conditional sampling distributions. See (22) for a formal definition of ALRErr_k,n_(· | ·). The accuracy of is almost indistinguishable from that of
, the most accurate of all approximate CSDs considered here. As expected, discretization reduces the accuracy somewhat, but even
is substantially more accurate than
and
. With θ0 = 0.01 and ρ0 = 0.05, we used the methodology described in the text to sample 250 conditional configurations, each with n = 10 haplotypes and k loci. (A) Error is measured relative to the true CSD π, estimated using computationally intensive importance sampling. (B) Error is measured relative to
, computed by numerically solving a recursion for the equivalent CSD
.
Figure 3.—
Absolute log-ratio error (ALRErr) of various conditional sampling distributions. See (22) for a formal definition of ALRErr_k,n_(· | ·). The accuracy of is almost indistinguishable from that of
, the most accurate of all approximate CSDs considered here. As expected, discretization reduces the accuracy somewhat, but even
is substantially more accurate than
and
. With θ0 = 0.01 and ρ0 = 0.05, we used the methodology described in the text to sample 250 conditional configurations, each with n = 10 haplotypes and k loci. (A) Error is measured relative to the true CSD π, estimated using computationally intensive importance sampling. (B) Error is measured relative to
, computed by numerically solving a recursion for the equivalent CSD
.
Figure 4.—
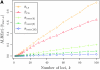
Comparison of the accuracy of various conditional sampling distributions relative to (see Figure 3 for the accuracy of
). A and B illustrate that the improvement in accuracy of
over
and
is amplified as the number of loci k increases and that both
and
produce significantly smaller values than
(and
). For θ0 = 0.01 and ρ0 = 0.05, we used the methodology described in the text to sample 250 conditional configurations with n = 10 haplotypes and k loci. (A) Absolute log-ratio error. (B) Signed log-ratio error.
Figure 4.—
Comparison of the accuracy of various conditional sampling distributions relative to (see Figure 3 for the accuracy of
). A and B illustrate that the improvement in accuracy of
over
and
is amplified as the number of loci k increases and that both
and
produce significantly smaller values than
(and
). For θ0 = 0.01 and ρ0 = 0.05, we used the methodology described in the text to sample 250 conditional configurations with n = 10 haplotypes and k loci. (A) Absolute log-ratio error. (B) Signed log-ratio error.
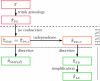
Figure 5.—
Illustration of the relationship between various CSDs. The CSD at the head of each arrow can be seen as an approximation to the CSD at the tail. Each arrow is also annotated with a (short) description of this approximation. The CSDs below the dashed line can be cast as an HMM: Those above the dotted line (including a continuous-state version of , which we denote
) have a continuous and infinite state space, while those below have a finite and discrete state space and are therefore amenable to simple dynamic programming algorithms. For more thorough descriptions of each approximation, see the main text and also P
aul
and S
ong
(2010). Recall in particular that the equality holds only for conditionally sampling a single haplotype.
Similar articles
- A sequentially Markov conditional sampling distribution for structured populations with migration and recombination.
Steinrücken M, Paul JS, Song YS. Steinrücken M, et al. Theor Popul Biol. 2013 Aug;87:51-61. doi: 10.1016/j.tpb.2012.08.004. Epub 2012 Sep 7. Theor Popul Biol. 2013. PMID: 23010245 Free PMC article. - A principled approach to deriving approximate conditional sampling distributions in population genetics models with recombination.
Paul JS, Song YS. Paul JS, et al. Genetics. 2010 Sep;186(1):321-38. doi: 10.1534/genetics.110.117986. Epub 2010 Jun 30. Genetics. 2010. PMID: 20592264 Free PMC article. - Estimating variable effective population sizes from multiple genomes: a sequentially markov conditional sampling distribution approach.
Sheehan S, Harris K, Song YS. Sheehan S, et al. Genetics. 2013 Jul;194(3):647-62. doi: 10.1534/genetics.112.149096. Epub 2013 Apr 22. Genetics. 2013. PMID: 23608192 Free PMC article. - Phase-type distributions in mathematical population genetics: An emerging framework.
Hobolth A, Rivas-González I, Bladt M, Futschik A. Hobolth A, et al. Theor Popul Biol. 2024 Jun;157:14-32. doi: 10.1016/j.tpb.2024.03.001. Epub 2024 Mar 7. Theor Popul Biol. 2024. PMID: 38460602 Review. - A practical introduction to sequentially Markovian coalescent methods for estimating demographic history from genomic data.
Mather N, Traves SM, Ho SYW. Mather N, et al. Ecol Evol. 2019 Dec 7;10(1):579-589. doi: 10.1002/ece3.5888. eCollection 2020 Jan. Ecol Evol. 2019. PMID: 31988743 Free PMC article. Review.
Cited by
- Exact Decoding of a Sequentially Markov Coalescent Model in Genetics.
Ki C, Terhorst J. Ki C, et al. J Am Stat Assoc. 2024;119(547):2242-2255. doi: 10.1080/01621459.2023.2252570. Epub 2023 Oct 3. J Am Stat Assoc. 2024. PMID: 39323740 Free PMC article. - A general and efficient representation of ancestral recombination graphs.
Wong Y, Ignatieva A, Koskela J, Gorjanc G, Wohns AW, Kelleher J. Wong Y, et al. Genetics. 2024 Sep 4;228(1):iyae100. doi: 10.1093/genetics/iyae100. Genetics. 2024. PMID: 39013109 Free PMC article. - A general and efficient representation of ancestral recombination graphs.
Wong Y, Ignatieva A, Koskela J, Gorjanc G, Wohns AW, Kelleher J. Wong Y, et al. bioRxiv [Preprint]. 2024 Apr 23:2023.11.03.565466. doi: 10.1101/2023.11.03.565466. bioRxiv. 2024. PMID: 37961279 Free PMC article. Updated. Preprint. - Properties of 2-locus genealogies and linkage disequilibrium in temporally structured samples.
Biddanda A, Steinrücken M, Novembre J. Biddanda A, et al. Genetics. 2022 May 5;221(1):iyac038. doi: 10.1093/genetics/iyac038. Genetics. 2022. PMID: 35294015 Free PMC article. - Variational inference using approximate likelihood under the coalescent with recombination.
Liu X, Ogilvie HA, Nakhleh L. Liu X, et al. Genome Res. 2021 Nov;31(11):2107-2119. doi: 10.1101/gr.273631.120. Epub 2021 Aug 23. Genome Res. 2021. PMID: 34426513 Free PMC article.
References
- Abramowitz, M., and I. A. Stegun (Editors), 1972. Handbook of Mathematical Functions With Formulas, Graphs, and Mathematical Tables. Dover, New York.
- Crawford, D. C., T. Bhangale, N. Li, G. Hellenthal, M. J. Rieder et al., 2004. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nat. Genet. 36 700–706. - PubMed
- De Iorio, M., and R. C. Griffiths, 2004. a Importance sampling on coalescent histories. I. Adv. Appl. Probab. 36(2): 417–433.
Publication types
MeSH terms
Grants and funding
- T32 HG000047/HG/NHGRI NIH HHS/United States
- K22 HG000047/HG/NHGRI NIH HHS/United States
- T32-HG00047/HG/NHGRI NIH HHS/United States
- R01 GM094402/GM/NIGMS NIH HHS/United States
- R01-GM094402/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources