Circadian clocks in human red blood cells - PubMed (original) (raw)
Circadian clocks in human red blood cells
John S O'Neill et al. Nature. 2011.
Abstract
Circadian (∼24 hour) clocks are fundamentally important for coordinated physiology in organisms as diverse as cyanobacteria and humans. All current models of the molecular circadian clockwork in eukaryotic cells are based on transcription-translation feedback loops. Non-transcriptional mechanisms in the clockwork have been difficult to study in mammalian systems. We circumvented these problems by developing novel assays using human red blood cells, which have no nucleus (or DNA) and therefore cannot perform transcription. Our results show that transcription is not required for circadian oscillations in humans, and that non-transcriptional events seem to be sufficient to sustain cellular circadian rhythms. Using red blood cells, we found that peroxiredoxins, highly conserved antioxidant proteins, undergo ∼24-hour redox cycles, which persist for many days under constant conditions (that is, in the absence of external cues). Moreover, these rhythms are entrainable (that is, tunable by environmental stimuli) and temperature-compensated, both key features of circadian rhythms. We anticipate that our findings will facilitate more sophisticated cellular clock models, highlighting the interdependency of transcriptional and non-transcriptional oscillations in potentially all eukaryotic cells.
Figures
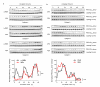
Figure 1. Circadian oscillation of peroxiredoxin (PRX) oxidation in human red blood cells
a, Red blood cells from three human subjects (A, B, C) were entrained by temperature cycles and then kept under constant conditions (at 37°C, in total darkness) and sampled every 4 hours. b, Red blood cells incubated in alternating 12 hour cycles of high (37°C) and low (32°C) temperature. Representative immunoblots showing oxidised/hyperoxidised peroxiredoxin (PRX-SO2/3) dimer are shown with loading controls. Quantification by densitometry is shown below. Values were normalised to the maximum for each blot. Solid line represents mean normalised intensity, with grey lines indicating s.e.m. boundaries. ** p < 0.01, *** p <0.001 by 1-way ANOVA (effect of time).
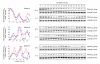
Figure 2. Circadian rhythms of peroxiredoxin (PRX) oxidation are not affected by transcriptional and translational inhibition
RBCs were entrained under temperature cycles and then kept under constant conditions (at 37°C, in total darkness) and sampled every 4 hours. Representative immunoblots showing oxidised/hyperoxidised peroxiredoxin (PRX-SO2/3) dimer are shown for samples incubated with a, α-amanitin (α-AMN), or b, cycloheximide (CHX) for the entirety of the experiments. Quantification by densitometry is shown below. Values were normalised to the maximum for each blot. Each point represents a mean normalised intensity. n.s., not significant. Further details are shown in Supplementary Fig. 3.
Figure 3. Temperature-compensation of circadian peroxiredoxin oxidation rhythms
Red blood cells were entrained in temperature cycles (12 h at 32°C, 12 h at 37°C) for two complete cycles and then kept under a constant temperature of either 32°C or 37°C for the rest of the experiment and sampled every 4 hours as before. Immunoblots for oxidised/hyperoxidised peroxiredoxin (PRX-SO2/3) dimer obtained from red blood cells from subjects A, B and C are shown. Loading controls (Coomassie-stained gels showing haemoglobin monomer bands) for each blot are also shown. Quantification of the above immunoblots by densitometry is shown on the left of the figure.
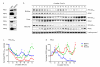
Figure 4. Expression patterns and oligomerisation of peroxiredoxins
a, Immunoblots showing expression of the human peroxiredoxin paralogues (PRX1-6) in red blood cells (RBC) and in mouse NIH3T3 cells. Loading of each lane was approximately equal. b, Oligomerisation patterns of PRX and PRX-SO2/3 in red blood cells. Following two cycles of temperature entrainment, cells were kept under constant temperature (37°C) for the rest of the experiment, and sampled every 4 hours. Representative immunoblots for PRX2 and PRX-SO2/3 are shown. Whole blot images in Supplementary Fig. 5 illustrate the different oligomeric forms. Immunoblots were quantified by densitometry for c, PRX-SO2/3 and d, PRX2. Arrowheads indicate peaks of abundance.
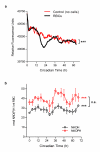
Figure 5. Circadian rhythms in haemoglobin oxidation and red blood cell (RBC) metabolism
a, Intrinsic front-face fluorescence (FFF) measurements of RBCs and controls. Experiments performed under constant conditions (at 37°C, in total darkness). Mean values for each time-point are shown (individual traces and further details are in Supplementary Fig. 6a). 2-way ANOVA (group × time) p < 0.001 (***). b, NADH and NADPH concentrations in red blood cells. Mean values (± s.e.m.) for three experimental subjects are shown. 1-way ANOVA (effect of time) for NADH/NADPH data was significant (*** p < 0.001). 2-way ANOVA (metabolite × time) did not reveal a significant difference between NADH and NADPH profiles (n.s., not significant). Individual profiles shown in Supplementary Fig. 6b,c.
Figure 6. Peroxiredoxin rhythms in nucleated cells
a, Peroxiredoxin rhythms in mouse NIH3T3 fibroblasts synchronised by a serum-shock. Immunoblots for Prx1, Prx6 and Prx-SO2/3 dimer are shown, in addition to Bmal1 and a β-actin loading control. b,c Peroxiredoxin rhythms in mouse embryonic fibroblasts (MEFs). MEFs from wild-type or mCry1/2 double-knockout mice were entrained in temperature cycles and then kept under constant temperature (37°C) for the rest of the experiment (as shown in the schematic). b, Representative immunoblots of oxidised/hyperoxidised peroxiredoxin (Prx-SO2/3) dimer. c, Quantification of Prx-SO2/3 immunoblots by densitometry. Mean values (± s.e.m.) for _n_=4 biological replicates are shown.
Comment in
- Circadian rhythms: Redox redux.
Bass J, Takahashi JS. Bass J, et al. Nature. 2011 Jan 27;469(7331):476-8. doi: 10.1038/469476a. Nature. 2011. PMID: 21270881 Free PMC article.
Similar articles
- Rhythmic glucose metabolism regulates the redox circadian clockwork in human red blood cells.
Ch R, Rey G, Ray S, Jha PK, Driscoll PC, Dos Santos MS, Malik DM, Lach R, Weljie AM, MacRae JI, Valekunja UK, Reddy AB. Ch R, et al. Nat Commun. 2021 Jan 15;12(1):377. doi: 10.1038/s41467-020-20479-4. Nat Commun. 2021. PMID: 33452240 Free PMC article. - Interplay between cellular redox oscillations and circadian clocks.
Rey G, Reddy AB. Rey G, et al. Diabetes Obes Metab. 2015 Sep;17 Suppl 1:55-64. doi: 10.1111/dom.12519. Diabetes Obes Metab. 2015. PMID: 26332969 Review. - Peroxiredoxins are conserved markers of circadian rhythms.
Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, Maywood ES, Hastings MH, Baliga NS, Merrow M, Millar AJ, Johnson CH, Kyriacou CP, O'Neill JS, Reddy AB. Edgar RS, et al. Nature. 2012 May 16;485(7399):459-64. doi: 10.1038/nature11088. Nature. 2012. PMID: 22622569 Free PMC article. - Circadian redox oscillations and metabolism.
Milev NB, Reddy AB. Milev NB, et al. Trends Endocrinol Metab. 2015 Aug;26(8):430-7. doi: 10.1016/j.tem.2015.05.012. Epub 2015 Jun 22. Trends Endocrinol Metab. 2015. PMID: 26113283 Free PMC article. Review. - Rethinking the clockwork: redox cycles and non-transcriptional control of circadian rhythms.
Wu L, Reddy AB. Wu L, et al. Biochem Soc Trans. 2014 Feb;42(1):1-10. doi: 10.1042/BST20130169. Biochem Soc Trans. 2014. PMID: 24450621
Cited by
- Circadian regulation of olfaction and an evolutionarily conserved, nontranscriptional marker in Caenorhabditis elegans.
Olmedo M, O'Neill JS, Edgar RS, Valekunja UK, Reddy AB, Merrow M. Olmedo M, et al. Proc Natl Acad Sci U S A. 2012 Dec 11;109(50):20479-84. doi: 10.1073/pnas.1211705109. Epub 2012 Nov 26. Proc Natl Acad Sci U S A. 2012. PMID: 23185015 Free PMC article. - Studying the Human Microbiota: Advances in Understanding the Fundamentals, Origin, and Evolution of Biological Timekeeping.
Siebieszuk A, Sejbuk M, Witkowska AM. Siebieszuk A, et al. Int J Mol Sci. 2023 Nov 10;24(22):16169. doi: 10.3390/ijms242216169. Int J Mol Sci. 2023. PMID: 38003359 Free PMC article. Review. - Transcriptome comparison between fetal and adult mouse livers: implications for circadian clock mechanisms.
Li C, Yu S, Zhong X, Wu J, Li X. Li C, et al. PLoS One. 2012;7(2):e31292. doi: 10.1371/journal.pone.0031292. Epub 2012 Feb 21. PLoS One. 2012. PMID: 22363607 Free PMC article. - Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression.
Kojima S, Sher-Chen EL, Green CB. Kojima S, et al. Genes Dev. 2012 Dec 15;26(24):2724-36. doi: 10.1101/gad.208306.112. Genes Dev. 2012. PMID: 23249735 Free PMC article. - The circadian clock gates the intestinal stem cell regenerative state.
Karpowicz P, Zhang Y, Hogenesch JB, Emery P, Perrimon N. Karpowicz P, et al. Cell Rep. 2013 Apr 25;3(4):996-1004. doi: 10.1016/j.celrep.2013.03.016. Epub 2013 Apr 11. Cell Rep. 2013. PMID: 23583176 Free PMC article.
References
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. - PubMed
- Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The adaptive value of circadian clocks; an experimental assessment in cyanobacteria. Curr Biol. 2004;14(16):1481–1486. - PubMed
- Dodd AN, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309(5734):630–633. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources