Removing noise from pyrosequenced amplicons - PubMed (original) (raw)
Removing noise from pyrosequenced amplicons
Christopher Quince et al. BMC Bioinformatics. 2011.
Abstract
Background: In many environmental genomics applications a homologous region of DNA from a diverse sample is first amplified by PCR and then sequenced. The next generation sequencing technology, 454 pyrosequencing, has allowed much larger read numbers from PCR amplicons than ever before. This has revolutionised the study of microbial diversity as it is now possible to sequence a substantial fraction of the 16S rRNA genes in a community. However, there is a growing realisation that because of the large read numbers and the lack of consensus sequences it is vital to distinguish noise from true sequence diversity in this data. Otherwise this leads to inflated estimates of the number of types or operational taxonomic units (OTUs) present. Three sources of error are important: sequencing error, PCR single base substitutions and PCR chimeras. We present AmpliconNoise, a development of the PyroNoise algorithm that is capable of separately removing 454 sequencing errors and PCR single base errors. We also introduce a novel chimera removal program, Perseus, that exploits the sequence abundances associated with pyrosequencing data. We use data sets where samples of known diversity have been amplified and sequenced to quantify the effect of each of the sources of error on OTU inflation and to validate these algorithms.
Results: AmpliconNoise outperforms alternative algorithms substantially reducing per base error rates for both the GS FLX and latest Titanium protocol. All three sources of error lead to inflation of diversity estimates. In particular, chimera formation has a hitherto unrealised importance which varies according to amplification protocol. We show that AmpliconNoise allows accurate estimates of OTU number. Just as importantly AmpliconNoise generates the right OTUs even at low sequence differences. We demonstrate that Perseus has very high sensitivity, able to find 99% of chimeras, which is critical when these are present at high frequencies.
Conclusions: AmpliconNoise followed by Perseus is a very effective pipeline for the removal of noise. In addition the principles behind the algorithms, the inference of true sequences using Expectation-Maximization (EM), and the treatment of chimera detection as a classification or 'supervised learning' problem, will be equally applicable to new sequencing technologies as they appear.
Figures
Figure 1
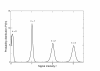
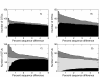
Flowgram signal intensity distributions. Probability distributions of observed signal intensities at different homopolymer lengths for the 'Even' V2 Mock Communities. The homopolymer length is shown above the mode of the distribution.
Figure 2
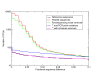
OTU numbers in the V5 'Artificial Community' as a function of percent sequence difference - logarithmic. Numbers of OTUs formed at cut-offs of increasing percent sequence difference after complete linkage clustering of the 'Artificial Community' V5 data set (Table 1). Distances were calculated following pair-wise alignment with the Needleman-Wunsch algorithm. Results are shown following filtering (red line), pyrosequencing noise removal by the first PyroNoise stage of AmpliconNoise (green line), further removal of PCR point mutations by the second SeqNoise stage (blue line) and following removal of chimeric sequences (magenta line). For comparison the number of OTUs obtained by clustering the known reference sequences are shown in black. The y-axis is logarithmically scaled.
Figure 3
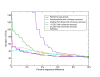
OTU numbers in the V5 'Artificial Community' as a function of percent sequence difference - linear. Numbers of OTUs formed at cut-offs of increasing percent sequence difference after complete linkage clustering of the 'Artificial Community' V5 data set (Table 1). Distances were calculated following pair-wise alignment with the Needleman-Wunsch algorithm, results are shown for the filtered sequences after pyrosequencing and PCR noise removal by AmpliconNoise (magenta line), for single-linkage preclustering at 1% (purple) and SLP at 2% (cyan), for the DeNoiser algorithm (orange), and for the original one-stage PyroNoise algorithm (dark green line). In all cases chimeric sequences were removed. For comparison the number of OTUs obtained by clustering the known reference sequences are shown in black. The y-axis is scaled linearly.
Figure 4
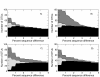
OTU construction accuracy for the V5 'Artificial Community' as a function of percent sequence difference for the different noise removal algorithms. Results are given for the improved two stage 'AmpliconNoise' (A), the original 'PyroNoise' (B), single-linkage preclustering at 2% (C), and the DeNoiser algorithm (D). Reads classified as chimeric by comparison with the references were removed. The solid black portion gives the number of OTUs comprised of reference sequences and denoised pyrosequenced reads. These are good OTUs. The grey area OTUs formed only from reference sequences. These correspond to true OTUs that are missed. The diagonal shaded area those OTUs containing only pyrosequenced reads and hence are noise.
Figure 5
OTU construction accuracy for the Titanium data set as a function of percent sequence difference for the different noise removal algorithms. Results are given for AmpliconNoise with σs = 0.01 (A) and σs = 0.04 (B), single-linkage preclustering at 2% (C), and the DeNoiser algorithm (D). Reads classified as chimeric by comparison with the references were removed. The solid black portion gives the number of OTUs comprised of reference sequences and denoised pyrosequenced reads. These are good OTUs. The grey area OTUs formed only from reference sequences. These correspond to true OTUs that are missed. The diagonal shaded area those OTUs containing only pyrosequenced reads and hence are noise.
Figure 6
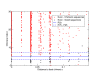
Training logistic regression on denoised V5 'Divergent' data. Good sequences are shown as black dots, chimeras red and reference sequences magenta. We used the denoised V5 'Divergent' data set, classified either good or chimeric by comparison with the references, and the reference sequences, all good, to train a one dimensional logistic regression on the 'chimera index' I using the R software package [30]. An intercept, α = - 183.25, and coefficient, β = 10.56, were obtained despite the fact that the algorithm did not converge (see text), and the corresponding P50 classification value, 17.35, is shown (blue line).
Figure 7
Validation of logistic regression on denoised V5 'Artificial Community' data. Applying the classification rule (blue line) from Figure 5 to the 'Artificial Community' denoised data sets correctly predicts all but two chimeras that fall below the P50 line. Good sequences are shown as blackdots and chimeras red.
Figure 8
Training logistic regression on denoised V2 'Even' data. Good sequences are shown as black dots, chimeras red and reference sequences magenta. We used the three denoised V2 'Even' data sets, classified either good or chimeric by comparison with the references, and the reference sequences, all good, to train a one dimensional logistic regression on the 'chimera index' I using the R software package. An intercept, α = - 2.83542, and coefficient β = 0.55889 were obtained (highly significantly different from zero). The corresponding P25, P50 and P75 decision lines are shown (blue lines). The fit reduced the null deviance from 1371.81 on 5416 degrees of freedom to a residual deviance of 416.36 on 5415 degrees of freedom (AIC 420.36).
References
- Margulies M, Egholm M, Altman W, Attiya S, Bader J, Bemben L, Berka J, Braverman M, Chen Y, Chen Z, Dewell S, Du L, Fierro J, Gomes X, Godwin B, He W, Helgesen S, Ho C, Irzyk G, Jando S, Alenquer M, Jarvie T, Jirage K, Kim J, Knight J, Lanza J, Leamon J, Lefkowitz S, Lei M, Li J, Lohman K, Lu H, Makhijani V, McDade K, McKenna M, Myers E, Nickerson E, Nobile J, Plant R, Puc B, Ronan M, Roth G, Sarkis G, Simons J, Simpson J, Srinivasan M, Tartaro K, Tomasz A, Vogt K, Volkmer G, Wang S, Wang Y, Weiner M, Yu P, Begley R, Rothberg J. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases