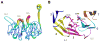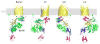The crystal structure of BamB suggests interactions with BamA and its role within the BAM complex - PubMed (original) (raw)
The crystal structure of BamB suggests interactions with BamA and its role within the BAM complex
Nicholas Noinaj et al. J Mol Biol. 2011.
Abstract
Escherichia coli BamB is the largest of four lipoproteins in the β-barrel assembly machinery (BAM) complex. It interacts with the periplasmic domain of BamA, an integral outer membrane protein (OMP) essential for OMP biogenesis. Although BamB is not essential, it serves an important function in the BAM complex, significantly increasing the folding efficiency of some OMPs in vivo and in vitro. To learn more about the BAM complex, we solved structures of BamB in three different crystal forms. BamB crystallized in space groups P2(1)3, I222, and P2(1)2(1)2(1), with one molecule per asymmetric unit in each case. Crystals from the space group I222 diffracted to 1. 65-Å resolution. BamB forms an eight-bladed β-propeller with a central pore and is shaped like a doughnut. A DALI search revealed that BamB shares structural homology to several eukaryotic proteins containing WD40 repeat domains, which commonly have β-propeller folds and often serve as scaffolding proteins within larger multi-protein complexes that carry out signal transduction, cell division, and chemotaxis. Using mutagenesis data from previous studies, we docked BamB onto a BamA structural model and assessed known and possible interactions between these two proteins. Our data suggest that BamB serves as a scaffolding protein within the BAM complex by optimally orienting the flexible periplasmic domain of BamA for interaction with other BAM components and chaperones. This may facilitate integration of newly synthesized OMPs into the outer membrane.
Published by Elsevier Ltd.
Figures
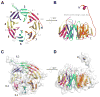
Figure 1
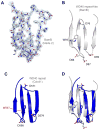
Overall structure of BamB. A, BamB has an eight bladed β-propeller fold with each blade shown in a different color (N-terminal residues 21–40 have been removed for clarity) and is rotated 90° about the x-axis in panel B. C, Space-filling representation of BamB rendered transparent and rotated 90° about the x-axis in panel D.
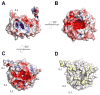
Figure 2
Surface characteristics of BamB. A, Electrostatic surface potential representation of BamB shows that the core region is strongly electronegative on both sides (B and C, rotated +90° and −90° about x-axis compared to panel A, respectively, to illustrate both sides). D. Surface hydrophobicity of BamB highlighting the most hydrophobic residues in yellow. The locations of IL2, IL4, and IL5 are indicated for reference.
Figure 3
BamB IL4 and IL5 are partially disordered and contain residues important for binding BamA. A, B-factor putty representation of BamB (P213) shows that the structure is mostly ordered with only N-terminal residues 21–40, IL4, and IL5 showing significant flexibility. B, Zoomed view indicating residues in BamB that were previously shown to lie within the binding interface with BamA. BamB is color coded according to Figure 1, important residues are shown in stick representation, and the disordered residues in IL5 are represented by a dashed line.
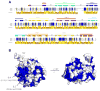
Figure 4
Sequence alignment of BamB homologs from Escherichia coli (EcBamB), Pseudomonas aeruginosa (PaBamB), and Vibrio cholerae (VcBamB). A, Residues having a conservation score of 7.5 or greater are color coded from yellow (7.5) to blue (11, max) and mapped onto the surface of the BamB structure. The BamB secondary structure is shown above the sequence alignment. Residues that have previously been reported to be involved in binding BamA are indicated by red asterisks. B, Conserved residues (blue) having a conservation score of 7.5 or greater were mapped onto the BamB crystal structure reported here.
Figure 5
BamB shows structural similarity to proteins containing WD40 repeat-like domains. A, β-propeller blade 2 from the BamB crystal structure (stick model) showing the electron density (blue mesh) from a σ– A weighted 2Fo-Fc map contoured at 1.0 σ. B, Ribbon representation of blade 2 illustrating its WD40 repeat-like domain structure, showing tryptophan and aspartate residues in stick representation. C, Ribbon representation of a WD40 repeat domain from Cdc41, showing tryptophan and aspartate residues in stick representation. D, Superposition of the WD40 domain from Cdc41 and the WD40 repeat-like domain from BamB.
Figure 6
Docking the BamA-BamB complex. A, Docked structure of BamB (green) onto POTRA 1-5, with each of the POTRA domains shown in a different color and in panel B, rotated 90° along the y-axis. C, Electrostatic charge distributions for POTRA 1-4, with BamB shown in green. D, Zoomed view of docked structure along POTRA 2-4, highlighting residues in IL4 (blue) and IL5 (pink) important for binding and a potential salt bridge formed between D241 (BamA) and R195 (BamB). As predicted from previous reports and obtained as a result in our docked model, strand β2 from POTRA 3 appears to interact with BamB IL4 (blue) by β-strand augmentation.
Figure 7
Model depicting how BamB interacts with BamA at the outer membrane. This model is based upon previously reported studies, homology to known structures, and our docking studies with the BamB structures.
Similar articles
- Crystal structure of BamB bound to a periplasmic domain fragment of BamA, the central component of the β-barrel assembly machine.
Jansen KB, Baker SL, Sousa MC. Jansen KB, et al. J Biol Chem. 2015 Jan 23;290(4):2126-36. doi: 10.1074/jbc.M114.584524. Epub 2014 Dec 2. J Biol Chem. 2015. PMID: 25468906 Free PMC article. - Crystal structure of Escherichia coli BamB, a lipoprotein component of the β-barrel assembly machinery complex.
Kim KH, Paetzel M. Kim KH, et al. J Mol Biol. 2011 Mar 11;406(5):667-78. doi: 10.1016/j.jmb.2010.12.020. Epub 2010 Dec 17. J Mol Biol. 2011. PMID: 21168416 - Structure of Escherichia coli BamB and its interaction with POTRA domains of BamA.
Dong C, Yang X, Hou HF, Shen YQ, Dong YH. Dong C, et al. Acta Crystallogr D Biol Crystallogr. 2012 Sep;68(Pt 9):1134-9. doi: 10.1107/S0907444912023141. Epub 2012 Aug 18. Acta Crystallogr D Biol Crystallogr. 2012. PMID: 22948914 - Augmenting β-augmentation: structural basis of how BamB binds BamA and may support folding of outer membrane proteins.
Heuck A, Schleiffer A, Clausen T. Heuck A, et al. J Mol Biol. 2011 Mar 11;406(5):659-66. doi: 10.1016/j.jmb.2011.01.002. Epub 2011 Jan 12. J Mol Biol. 2011. PMID: 21236263 Review. - The β-Barrel Assembly Machinery Complex.
Leyton DL, Belousoff MJ, Lithgow T. Leyton DL, et al. Methods Mol Biol. 2015;1329:1-16. doi: 10.1007/978-1-4939-2871-2_1. Methods Mol Biol. 2015. PMID: 26427672 Review.
Cited by
- BB0324 and BB0028 are constituents of the Borrelia burgdorferi β-barrel assembly machine (BAM) complex.
Lenhart TR, Kenedy MR, Yang X, Pal U, Akins DR. Lenhart TR, et al. BMC Microbiol. 2012 Apr 20;12:60. doi: 10.1186/1471-2180-12-60. BMC Microbiol. 2012. PMID: 22519960 Free PMC article. - Genetic, biochemical, and molecular characterization of the polypeptide transport-associated domain of Escherichia coli BamA.
Workman P, Heide K, Giuliano N, Lee N, Mar J, Vuong P, Bennion D, Misra R. Workman P, et al. J Bacteriol. 2012 Jul;194(13):3512-21. doi: 10.1128/JB.06740-11. Epub 2012 Apr 27. J Bacteriol. 2012. PMID: 22544271 Free PMC article. - The structural biology of β-barrel membrane proteins: a summary of recent reports.
Fairman JW, Noinaj N, Buchanan SK. Fairman JW, et al. Curr Opin Struct Biol. 2011 Aug;21(4):523-31. doi: 10.1016/j.sbi.2011.05.005. Epub 2011 Jun 28. Curr Opin Struct Biol. 2011. PMID: 21719274 Free PMC article. Review. - Analysis of the binding forces driving the tight interactions between beta-lactamase inhibitory protein-II (BLIP-II) and class A beta-lactamases.
Brown NG, Chow DC, Sankaran B, Zwart P, Prasad BV, Palzkill T. Brown NG, et al. J Biol Chem. 2011 Sep 16;286(37):32723-35. doi: 10.1074/jbc.M111.265058. Epub 2011 Jul 20. J Biol Chem. 2011. PMID: 21775426 Free PMC article. - Crystal structure of BamB bound to a periplasmic domain fragment of BamA, the central component of the β-barrel assembly machine.
Jansen KB, Baker SL, Sousa MC. Jansen KB, et al. J Biol Chem. 2015 Jan 23;290(4):2126-36. doi: 10.1074/jbc.M114.584524. Epub 2014 Dec 2. J Biol Chem. 2015. PMID: 25468906 Free PMC article.
References
- Knowles TJ, Scott-Tucker A, Overduin M, Henderson IR. Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat Rev Microbiol. 2009;7:206–14. - PubMed
- Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–45. - PubMed
- Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, Misra R, Silhavy TJ. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol. 2006;61:151–64. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous