PhylOTU: a high-throughput procedure quantifies microbial community diversity and resolves novel taxa from metagenomic data - PubMed (original) (raw)
PhylOTU: a high-throughput procedure quantifies microbial community diversity and resolves novel taxa from metagenomic data
Thomas J Sharpton et al. PLoS Comput Biol. 2011.
Abstract
Microbial diversity is typically characterized by clustering ribosomal RNA (SSU-rRNA) sequences into operational taxonomic units (OTUs). Targeted sequencing of environmental SSU-rRNA markers via PCR may fail to detect OTUs due to biases in priming and amplification. Analysis of shotgun sequenced environmental DNA, known as metagenomics, avoids amplification bias but generates fragmentary, non-overlapping sequence reads that cannot be clustered by existing OTU-finding methods. To circumvent these limitations, we developed PhylOTU, a computational workflow that identifies OTUs from metagenomic SSU-rRNA sequence data through the use of phylogenetic principles and probabilistic sequence profiles. Using simulated metagenomic data, we quantified the accuracy with which PhylOTU clusters reads into OTUs. Comparisons of PCR and shotgun sequenced SSU-rRNA markers derived from the global open ocean revealed that while PCR libraries identify more OTUs per sequenced residue, metagenomic libraries recover a greater taxonomic diversity of OTUs. In addition, we discover novel species, genera and families in the metagenomic libraries, including OTUs from phyla missed by analysis of PCR sequences. Taken together, these results suggest that PhylOTU enables characterization of part of the biosphere currently hidden from PCR-based surveys of diversity?
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
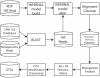
Figure 1. PhylOTU Workflow.
Computational processes are represented as squares and databases are represented as cylinders in this generalize workflow of
PhylOTU
. See Results section for details.
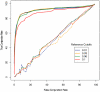
Figure 2. Relationship between false clustering rate and true clustering rate.
Each read data set was clustered into OTUs at various thresholds and compared to the corresponding full-length data set, which was clustered at several fixed PD thresholds (shown here are full-length sequence cutoffs of 0.01, 0.03, 0.05 and 0.1). For each full-length sequence threshold, the true conjunction and false conjunction rates of the read OTUs were calculated as a function of the read threshold. Solid lines represent the median value of the true and false conjunction rates across simulations. Dashed lines represent the median value of the true and false conjunction rates derived from comparisons of randomly permuted clusters relative to the source sequence clusters.
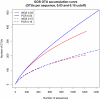
Figure 3. Rarefaction analysis of OTUs identified from PCR and metagenomic sequencing at two different sequence similarity cutoffs (solid = 0.03, dashed = 0.15).
Rarefaction curves are shown for OTUs from PCR (blue) and metagenomic (red) sequencing libraries. Two different sequence similarity cutoffs are used (solid = 0.03, dashed = 0.15). Curves represent the average number of OTUs per sequence from 100 random draws of subsets of sequences from each data set.
Figure 4. Overlap between GOS OTUs revealed by PCR sequencing and metagenomic sequencing.
PhylOTU
was used to identify OTUs from a data set comprised of both PCR and shotgun SSU-rRNA sequences obtained from six Global Ocean Survey samples. The 1,254 OTUs that contained only PCR sequences at a clustering threshold of 0.03 were designated as OTUs unique to PCR, while those 80 OTUs that contained only metagenomic sequences at the corresponding clustering threshold of 0.15 (see Results) were designated as OTUs unique to metagenomic sequencing. The 309 OTUs identified by both PCR and shotgun sequencing was determined by using a clustering threshold of 0.03 (162 shared OTUs are identified when a threshold of 0.15 is used). The total number of OTUs and the total number of SSU-rRNA sequences are shown on the left and right of the Venn-Diagram for the PCR (threshold = 0.03) and metagenomic data (threshold = 0.15), respectively. The taxonomic distribution of each set of OTUs is shown beneath the Venn-Diagram. Here, every sequence from each OTU was taxonomically classified into major clades of Bacteria (approximately phylum level designations) using the Ribosomal Database Project classification software. The relative abundance of each taxonomic group is plotted along the x-axis (specific values can be found in Table S5). Clades exhibiting less than 1% relative abundance across all sets of OTUs are not shown.
Similar articles
- The phylogenetic diversity of metagenomes.
Kembel SW, Eisen JA, Pollard KS, Green JL. Kembel SW, et al. PLoS One. 2011;6(8):e23214. doi: 10.1371/journal.pone.0023214. Epub 2011 Aug 31. PLoS One. 2011. PMID: 21912589 Free PMC article. - Groundtruthing next-gen sequencing for microbial ecology-biases and errors in community structure estimates from PCR amplicon pyrosequencing.
Lee CK, Herbold CW, Polson SW, Wommack KE, Williamson SJ, McDonald IR, Cary SC. Lee CK, et al. PLoS One. 2012;7(9):e44224. doi: 10.1371/journal.pone.0044224. Epub 2012 Sep 6. PLoS One. 2012. PMID: 22970184 Free PMC article. - DBH: A de Bruijn graph-based heuristic method for clustering large-scale 16S rRNA sequences into OTUs.
Wei ZG, Zhang SW. Wei ZG, et al. J Theor Biol. 2017 Jul 21;425:80-87. doi: 10.1016/j.jtbi.2017.04.019. Epub 2017 Apr 26. J Theor Biol. 2017. PMID: 28454900 - Ecological consistency of SSU rRNA-based operational taxonomic units at a global scale.
Schmidt TS, Matias Rodrigues JF, von Mering C. Schmidt TS, et al. PLoS Comput Biol. 2014 Apr 24;10(4):e1003594. doi: 10.1371/journal.pcbi.1003594. eCollection 2014 Apr. PLoS Comput Biol. 2014. PMID: 24763141 Free PMC article. - Metagenomic Analysis Using Phylogenetic Placement-A Review of the First Decade.
Czech L, Stamatakis A, Dunthorn M, Barbera P. Czech L, et al. Front Bioinform. 2022 May 26;2:871393. doi: 10.3389/fbinf.2022.871393. eCollection 2022. Front Bioinform. 2022. PMID: 36304302 Free PMC article. Review.
Cited by
- Experimental and analytical tools for studying the human microbiome.
Kuczynski J, Lauber CL, Walters WA, Parfrey LW, Clemente JC, Gevers D, Knight R. Kuczynski J, et al. Nat Rev Genet. 2011 Dec 16;13(1):47-58. doi: 10.1038/nrg3129. Nat Rev Genet. 2011. PMID: 22179717 Free PMC article. Review. - Identification of rare microbial colonizers of plastic materials incubated in a coral reef environment.
Singleton SL, Davis EW 2nd, Arnold HK, Daniels AMY, Brander SM, Parsons RJ, Sharpton TJ, Giovannoni SJ. Singleton SL, et al. Front Microbiol. 2023 Oct 5;14:1259014. doi: 10.3389/fmicb.2023.1259014. eCollection 2023. Front Microbiol. 2023. PMID: 37869676 Free PMC article. - Phylogeny of bacterial and archaeal genomes using conserved genes: supertrees and supermatrices.
Lang JM, Darling AE, Eisen JA. Lang JM, et al. PLoS One. 2013 Apr 25;8(4):e62510. doi: 10.1371/journal.pone.0062510. Print 2013. PLoS One. 2013. PMID: 23638103 Free PMC article. - Direct 16S rRNA-seq from bacterial communities: a PCR-independent approach to simultaneously assess microbial diversity and functional activity potential of each taxon.
Rosselli R, Romoli O, Vitulo N, Vezzi A, Campanaro S, de Pascale F, Schiavon R, Tiarca M, Poletto F, Concheri G, Valle G, Squartini A. Rosselli R, et al. Sci Rep. 2016 Aug 31;6:32165. doi: 10.1038/srep32165. Sci Rep. 2016. PMID: 27577787 Free PMC article. - Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics.
Poretsky R, Rodriguez-R LM, Luo C, Tsementzi D, Konstantinidis KT. Poretsky R, et al. PLoS One. 2014 Apr 8;9(4):e93827. doi: 10.1371/journal.pone.0093827. eCollection 2014. PLoS One. 2014. PMID: 24714158 Free PMC article.
References
- Martiny JH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, et al. Microbial biogeography: putting microorganisms on the map. Nat Rev Micro. 2006;4:102–12. - PubMed