The biophysical and molecular basis of TRPV1 proton gating - PubMed (original) (raw)
The biophysical and molecular basis of TRPV1 proton gating
Eduardo Aneiros et al. EMBO J. 2011.
Abstract
The capsaicin receptor TRPV1, a member of the transient receptor potential family of non-selective cation channels is a polymodal nociceptor. Noxious thermal stimuli, protons, and the alkaloid irritant capsaicin open the channel. The mechanisms of heat and capsaicin activation have been linked to voltage-dependent gating in TRPV1. However, until now it was unclear whether proton activation or potentiation or both are linked to a similar voltage-dependent mechanism and which molecular determinants underlie the proton gating. Using the whole-cell patch-clamp technique, we show that protons activate and potentiate TRPV1 by shifting the voltage dependence of the activation curves towards more physiological membrane potentials. We further identified a key residue within the pore region of TRPV1, F660, to be critical for voltage-dependent proton activation and potentiation. We conclude that proton activation and potentiation of TRPV1 are both voltage dependent and that amino acid 660 is essential for proton-mediated gating of TRPV1.
Conflict of interest statement
The authors declare that they have no conflict of interest. All authors are Pfizer employees.
Figures
Figure 1
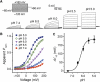
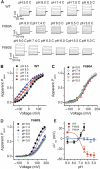
Voltage shift of activation curve of TRPV1 by protons. (A) Representative whole-cell current traces elicited by a voltage step protocol in response to protons as indicated. (B) Normalized steady-state maximal conductance curves at different pH values obtained from current traces shown in A. Lines represent Boltzmann fit to the data. (C) Δ_V_1/2 as a function of pH (_n_=7). Solid line represents Hill fit to the data. Error bars represent s.e.m.
Figure 2
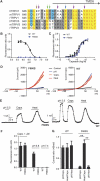
TRPV1 mutant F660S ablates proton activation but not capsaicin or heat activation. (A)An amino-acid sequence alignment of part of the pore region, including the upper third of TMD6 of human (h), mouse (m), and rat (r) TRPV1 and TRPV3. Residues reported to be specifically required for temperature activation of TRPV3 are indicated by purple arrows (I644, P651, L657, and Y661). Amino acids found to be required for temperature activation of both rTRPV1 and mTRPV3 are indicated by green arrows. The blue arrow indicates amino acid involved in heat activation of mTRPV3 and proton activation and potentiation of hTRPV1. The brown arrow indicates amino acid involved in heat activation of mTRPV3 and proton activation of hTRPV1. (B, C) Concentration response curves obtained from Fluo-4-based calcium flux experiments using HEK293 cells transiently expressing hTRPV1 wild-type or TRPV1 (F660S) mutant. Representative pH dose–response measurement (B) using MES-buffered solutions adjusted to different pH values. MES-buffered solutions were added to cells loaded with Fluo-4 calcium indicator dye in standard buffer solution without HEPES (adjusted to pH 7.4) resulting in final pH values of 5.6, 5.8, 6.2, 6.6, 7.0, and 7.4. Representative capsaicin dose–response measurement (C) performed in standard buffer solution containing 20 mM HEPES, adjusted to pH 7.4. Average EC50 values (mean±s.d.) of 25±12 nM were measured for F660S versus 12±6 nM for wild-type TRPV1 (_n_=10, each). (D) Steady-state current–voltage relationships obtained from whole-cell patch-clamp experiments with HEK293 cells transiently expressing hTRPV1 wild-type or F660S mutant. Cells were stimulated subsequently with protons (pH 5.5), capsaicin (1 μM), or heat (45°C). (E) Representative whole-cell current traces (at +60 mV) obtained from HEK293 cells transiently expressing hTRPV1 or F660S mutant after subsequent stimulation with protons (pH 5.5), capsaicin (100 nM), or heat (45°C). (F) Bar diagram summarizing data obtained from experiments as described in B and C. (G) Bar diagram showing ratio values calculated based on data as described in D and E. Statistical analysis was performed using one-way ANOVA followed by Tukey's post test; **P<0.001.
Figure 3
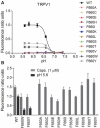
An aromatic amino acid at position 660 is required for proton activation. (A) Calcium flux experiments as described in Figure 2B, showing pH concentration response curves obtained from HEK293 cells transiently transfected with either hTRPV1 wild type or one of the following mutants: F660A, F660C, F660E, F660H, F660I, F660K, F660L, F660S, F660T, F660V, F660W, and F660Y. Only wild type, F660Y, and F660W responded to proton stimulation. (B) Bar diagram summarizing results as shown in A, obtained from at least five independent experiments, each.
Figure 4
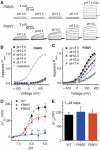
Mutations at position 660 affect the voltage dependence of proton activation. (A) Representative whole-cell current traces for TRPV1 (F660S) and TRPV1 (F660Y) mutants elicited by a voltage step protocol in response to protons as indicated. (B, C) Normalized steady-state maximal conductance curves at different pH values obtained from current traces shown in A. Lines represent Boltzmann fit to the data. (D) Δ_V_1/2 as a function of pH for wild type, F660S, and F660Y (_n_=5–6, *P<0.05, **_P_<0.01). Lines represent Hill fit to the data. Error bars represent s.e.m. (**E**) Comparison of Δ_V_1/2 for wild type, F660S, and F660Y in response to 1 μM capsaicin (_n_=4–5, _P_>0.05). Error bars represent s.e.m.
Figure 5
Proton potentiation is affected in TRPV1 (F660) mutants. (A–C) Effect of acidic pH on capsaicin stimulation in HEK293 cells expressing either hTRPV1 wild-type (A), TRPV1 (F660S), or TRPV1(F660A) mutant (B, C). Shown are representative current traces (at +60 mV and −60 mV), each. Substitution of F660 to alanine caused a selective loss of activation and potentiation by protons (pH 5.5) but did not cause inhibition of capsaicin responses as shown for F660S. Substitution of F660 to tyrosine did not result in a complete loss of activation by protons, but caused an inhibition of capsaicin responses similar to F660S. (D) Summary of data obtained from experiments as shown in (A–C). All values are shown as mean±s.e.m.
Figure 6
Proton-mediated sensitization of TRPV1 capsaicin activation is voltage dependent. (A) Representative whole-cell current traces of wild-type and mutant TRPV1 (F660A and F660S) elicited by 100 nM capsaicin at different pH values as indicated. C means the indicated pH solution contained 100 nM capsaicin. (B–D) Normalized steady-state maximal conductance curves at different pH values obtained from current traces shown in A. Lines represent Boltzmann fit to the data. (E) Δ_V_1/2 as a function of pH for wild type, F660A and F660S currents elicited by 100 nM capsaicin (_n_=5–6). Lines represent Hill fit to wild type and F660S and linear fit to F660A data. Error bars represent s.e.m.
Figure 7
Summary of known and novel pH mutants of TRPV1. (A) Schematic topology showing the putative positions of TRPV1 point mutations involved in proton activation or potentiation (brown) or activation and potentiation (blue). Letter(s) in parentheses indicate whether the respective position is in human (h) and/or rat (r) TRPV1. (B) Published and novel TRPV1 point mutants with effect on proton activation and/or potentiation of TRPV1.
Similar articles
- Protons stabilize the closed conformation of gain-of-function mutants of the TRPV1 channel.
Boukalova S, Teisinger J, Vlachova V. Boukalova S, et al. Biochim Biophys Acta. 2013 Mar;1833(3):520-8. doi: 10.1016/j.bbamcr.2012.11.017. Epub 2012 Dec 4. Biochim Biophys Acta. 2013. PMID: 23220012 - Temperature-induced opening of TRPV1 ion channel is stabilized by the pore domain.
Grandl J, Kim SE, Uzzell V, Bursulaya B, Petrus M, Bandell M, Patapoutian A. Grandl J, et al. Nat Neurosci. 2010 Jun;13(6):708-14. doi: 10.1038/nn.2552. Epub 2010 Apr 22. Nat Neurosci. 2010. PMID: 20414199 Free PMC article. - TRPV1 pore turret dictates distinct DkTx and capsaicin gating.
Geron M, Kumar R, Zhou W, Faraldo-Gómez JD, Vásquez V, Priel A. Geron M, et al. Proc Natl Acad Sci U S A. 2018 Dec 11;115(50):E11837-E11846. doi: 10.1073/pnas.1809662115. Epub 2018 Nov 21. Proc Natl Acad Sci U S A. 2018. PMID: 30463948 Free PMC article. - Differential effects of TRPV channel block on polymodal activation of rat cutaneous nociceptors in vitro.
St Pierre M, Reeh PW, Zimmermann K. St Pierre M, et al. Exp Brain Res. 2009 Jun;196(1):31-44. doi: 10.1007/s00221-009-1808-3. Epub 2009 Apr 30. Exp Brain Res. 2009. PMID: 19404626 Review. - Capsaicin receptor: TRPV1 a promiscuous TRP channel.
Pingle SC, Matta JA, Ahern GP. Pingle SC, et al. Handb Exp Pharmacol. 2007;(179):155-71. doi: 10.1007/978-3-540-34891-7_9. Handb Exp Pharmacol. 2007. PMID: 17217056 Review.
Cited by
- Molecular mechanism of TRP channels.
Zheng J. Zheng J. Compr Physiol. 2013 Jan;3(1):221-42. doi: 10.1002/cphy.c120001. Compr Physiol. 2013. PMID: 23720286 Free PMC article. Review. - Single residues in the outer pore of TRPV1 and TRPV3 have temperature-dependent conformations.
Kim SE, Patapoutian A, Grandl J. Kim SE, et al. PLoS One. 2013;8(3):e59593. doi: 10.1371/journal.pone.0059593. Epub 2013 Mar 26. PLoS One. 2013. PMID: 23555720 Free PMC article. - Harnessing the Therapeutic Potential of Capsaicin and Its Analogues in Pain and Other Diseases.
Basith S, Cui M, Hong S, Choi S. Basith S, et al. Molecules. 2016 Jul 23;21(8):966. doi: 10.3390/molecules21080966. Molecules. 2016. PMID: 27455231 Free PMC article. Review. - SUMOylation and DeSUMOylation: Tug of War of Pain Signaling.
Calderon-Rivera A, Gomez K, Rodríguez-Palma EJ, Khanna R. Calderon-Rivera A, et al. Mol Neurobiol. 2024 Sep 14. doi: 10.1007/s12035-024-04478-w. Online ahead of print. Mol Neurobiol. 2024. PMID: 39276308 Review. - Differential effects of acute hypoxia on the activation of TRPV1 by capsaicin and acidic pH.
Kim KS, Yoo HY, Park KS, Kim JK, Zhang YH, Kim SJ. Kim KS, et al. J Physiol Sci. 2012 Mar;62(2):93-103. doi: 10.1007/s12576-011-0185-4. Epub 2012 Jan 4. J Physiol Sci. 2012. PMID: 22215506 Free PMC article.
References
- Chou MZ, Mtui T, Gao YD, Kohler M, Middleton RE (2004) Resiniferatoxin binds to the capsaicin receptor (TRPV1) near the extracellular side of the S4 transmembrane domain. Biochemistry 43: 2501–2511 - PubMed
- Clapham DE (2003) TRP channels as cellular sensors. Nature 426: 517–524 - PubMed
- Clapham DE, Montell C, Schultz G, Julius D (2003) International Union of Pharmacology. XLIII. Compendium of voltage-gated ion channels: transient receptor potential channels. Pharmacol Rev 55: 591–596 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources