Mapping copy number variation by population-scale genome sequencing - PubMed (original) (raw)
. 2011 Feb 3;470(7332):59-65.
doi: 10.1038/nature09708.
Klaudia Walter, Chip Stewart, Robert E Handsaker, Ken Chen, Can Alkan, Alexej Abyzov, Seungtai Chris Yoon, Kai Ye, R Keira Cheetham, Asif Chinwalla, Donald F Conrad, Yutao Fu, Fabian Grubert, Iman Hajirasouliha, Fereydoun Hormozdiari, Lilia M Iakoucheva, Zamin Iqbal, Shuli Kang, Jeffrey M Kidd, Miriam K Konkel, Joshua Korn, Ekta Khurana, Deniz Kural, Hugo Y K Lam, Jing Leng, Ruiqiang Li, Yingrui Li, Chang-Yun Lin, Ruibang Luo, Xinmeng Jasmine Mu, James Nemesh, Heather E Peckham, Tobias Rausch, Aylwyn Scally, Xinghua Shi, Michael P Stromberg, Adrian M Stütz, Alexander Eckehart Urban, Jerilyn A Walker, Jiantao Wu, Yujun Zhang, Zhengdong D Zhang, Mark A Batzer, Li Ding, Gabor T Marth, Gil McVean, Jonathan Sebat, Michael Snyder, Jun Wang, Kenny Ye, Evan E Eichler, Mark B Gerstein, Matthew E Hurles, Charles Lee, Steven A McCarroll, Jan O Korbel; 1000 Genomes Project
Collaborators, Affiliations
- PMID: 21293372
- PMCID: PMC3077050
- DOI: 10.1038/nature09708
Mapping copy number variation by population-scale genome sequencing
Ryan E Mills et al. Nature. 2011.
Abstract
Genomic structural variants (SVs) are abundant in humans, differing from other forms of variation in extent, origin and functional impact. Despite progress in SV characterization, the nucleotide resolution architecture of most SVs remains unknown. We constructed a map of unbalanced SVs (that is, copy number variants) based on whole genome DNA sequencing data from 185 human genomes, integrating evidence from complementary SV discovery approaches with extensive experimental validations. Our map encompassed 22,025 deletions and 6,000 additional SVs, including insertions and tandem duplications. Most SVs (53%) were mapped to nucleotide resolution, which facilitated analysing their origin and functional impact. We examined numerous whole and partial gene deletions with a genotyping approach and observed a depletion of gene disruptions amongst high frequency deletions. Furthermore, we observed differences in the size spectra of SVs originating from distinct formation mechanisms, and constructed a map of SV hotspots formed by common mechanisms. Our analytical framework and SV map serves as a resource for sequencing-based association studies.
Figures
Figure 1. SV discovery and genotyping in population scale sequence data
A. Schematic depicting the different modes (i.e., approaches) of sequence based SV detection we used. The RP approach assesses the orientation and spacing of the mapped reads of paired-end sequences, (reads are denoted by arrows); the RD approach evaluates the read depth-of-coverage,; the SR approach maps the boundaries (breakpoints) of SVs by sequence alignment,; the AS approach assembles SVs,,. B. Integrated pipeline for SV discovery, validation, and genotyping. Colored circles represent individual SV discovery methods (listed in Supplementary Table 1), with modes indicated by a color scheme: green=RP; yellow=RD; purple=SR; red=AS; green and yellow=methods evaluating RP and RD (abbreviated as ‘PD’). C. Example of a deletion, previously associated with BMI, identified independently with RP (green), RD (yellow), and SR (red) methods. Grey dots indicate position and mapping quality for individual sequence reads. Targeted assembly confirmed the breakpoints detected by SR.
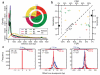
Figure 2. Comparative assessment of deletion discovery methods
A. Deletion size-range ascertained by different modes of SV discovery. Three groups are visible, with AS and SR, PD and RP, as well as RD and ‘RL’ (RP analysis involving relatively long range (≥1 kb) insert size libraries, resulting in a different deletion detection size range compared to the predominantly used <500kb insert size libraries), respectively, ascertaining similar size-ranges. Pie charts display the contribution of different SV discovery modes to the release set. Outer pie = based on number of SV calls; inner pie = based on total number of variable nucleotides. Of note, not all approaches were applied across all individuals (see Supplementary Table 2). B. Sensitivity and FDR estimates for individual deletion discovery methods based on gold standard sets for individuals sequenced at high (NA12878) and low-coverage (NA12156), respectively. All depicted estimates are summarized in Supplementary Tables 3, 4, 6. Vertical dotted lines correspond to the specificity threshold (FDR≤10%). C. Breakpoint mapping resolution of three deletion discovery methods (the respective method names are in Supplementary Table 2). The blue and red histograms are the breakpoint residuals for predicted deletion start and end coordinates, respectively, relative to assembled coordinates (here assessed in low-coverage data). The horizontal lines at the top of each plot mark the 98% confidence intervals (labeled for each panel), with vertical notches indicating the positions of the most probable breakpoint (the distribution mode).
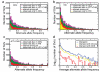
Figure 3. Analysis of deletion presence and absence in two populations
A-C. Deletion allele frequencies and observed sharing of alleles across populations, displayed for deletions discovered in the CEU, YRI, and JPT+CHB population samples in terms of stacked bars. D. Allele frequency spectra for deletions intersecting with intergenic (blue), intronic (yellow), and protein-coding sequences (red).
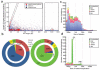
Figure 4. Contribution of SV formation mechanisms to the SV size spectrum
A. Breakpoint junction homology/microhomology length plotted as a function of SV size for SVs originally identified as deletions compared to a human reference. Dots are colored according to the SVs’ classification as deletions, insertions/duplications, or “undetermined” relative to inferred ancestral genomic loci. Gray lines mark groups of SVs likely formed by a common formation mechanism. The diagonal highlights tandem duplications (and few reciprocal deletion events), in which the length of the duplicated sequence correlates linearly with the length of the longest breakpoint junction sequence identity stretch. The ellipses indicate MEIs, i.e., Alu (~300 bp) and L1 (~6 kb) insertions, associated with target site duplications of up to 28 bp in size at the breakpoints. The horizontal group corresponds mostly to NH-associated deletions with <10 bp microhomology at the breakpoints. The remaining (ungrouped) SVs comprise truncated MEIs, VNTR expansion and shrinkage events, as well as NAHR-associated deletions and duplications. B. Relative contributions of SV formation mechanisms in the genome. Numbers of SVs are displayed on the outer pie chart and affected base pairs on the inner. Left panel: SVs classified as deletions relative to ancestral loci. Right panel: SVs classified as insertions/duplications. C. Size spectra of deletions classified relative to ancestral loci. D. Size spectra of insertions/duplications.
Figure 5. Mapping hotspots of SV formation in the genome
A. Distribution of SVs on chromosome 10 (“chr10”). Above the ideogram, colored bars indicate SV formation mechanisms (same color scheme as in B and C); bar lengths relate to the logarithm of SV size. Below the ideogram, bar lengths are directly proportional to allele frequencies. Arrows indicate an SV hotspot near the centromere underlying mainly VNTR, and several hotspots near the telomeres underlying mainly NAHR events. B. Enrichment of SVs inferred to be formed by the same formation mechanism for different genomic window sizes. Displayed is an enrichment of nearby, non-overlapping SVs formed by the same mechanism relative to an SV set where mechanism assignments are shuffled randomly. C. SV hotspots are mostly dominated by a single formation mechanism. Colored bars depict numbers of SV hotspots in which at least 50% of the variants were inferred to be formed by a single formation mechanism. The average abundance of NAHR-classified SVs in NAHR hotspots was 70% (compared with 77% for VNTR-hotspots; 69% for NH). The gray bar (“mixed”) corresponds to SV hotspots with no single mechanism dominating.
Similar articles
- High-resolution structural variants catalogue in a large-scale whole genome sequenced bovine family cohort data.
Lee YL, Bosse M, Takeda H, Moreira GCM, Karim L, Druet T, Oget-Ebrad C, Coppieters W, Veerkamp RF, Groenen MAM, Georges M, Bouwman AC, Charlier C. Lee YL, et al. BMC Genomics. 2023 May 1;24(1):225. doi: 10.1186/s12864-023-09259-8. BMC Genomics. 2023. PMID: 37127590 Free PMC article. - The fine-scale architecture of structural variants in 17 mouse genomes.
Yalcin B, Wong K, Bhomra A, Goodson M, Keane TM, Adams DJ, Flint J. Yalcin B, et al. Genome Biol. 2012;13(3):R18. doi: 10.1186/gb-2012-13-3-r18. Genome Biol. 2012. PMID: 22439878 Free PMC article. - Impact of genomic structural variation in Drosophila melanogaster based on population-scale sequencing.
Zichner T, Garfield DA, Rausch T, Stütz AM, Cannavó E, Braun M, Furlong EE, Korbel JO. Zichner T, et al. Genome Res. 2013 Mar;23(3):568-79. doi: 10.1101/gr.142646.112. Epub 2012 Dec 6. Genome Res. 2013. PMID: 23222910 Free PMC article. - Genome structural variation in human evolution.
Hollox EJ, Zuccherato LW, Tucci S. Hollox EJ, et al. Trends Genet. 2022 Jan;38(1):45-58. doi: 10.1016/j.tig.2021.06.015. Epub 2021 Jul 17. Trends Genet. 2022. PMID: 34284881 Review. - A Practical Guide for Structural Variation Detection in the Human Genome.
Yang L. Yang L. Curr Protoc Hum Genet. 2020 Sep;107(1):e103. doi: 10.1002/cphg.103. Curr Protoc Hum Genet. 2020. PMID: 32813322 Free PMC article. Review.
Cited by
- Analysis of genomic copy number variations through whole-genome scan in Yunling cattle.
Dang D, Zhang L, Gao L, Peng L, Chen J, Yang L. Dang D, et al. Front Vet Sci. 2024 Jul 22;11:1413504. doi: 10.3389/fvets.2024.1413504. eCollection 2024. Front Vet Sci. 2024. PMID: 39104544 Free PMC article. - Genome-wide detection of copy number variation in American mink using whole-genome sequencing.
Davoudi P, Do DN, Rathgeber B, Colombo SM, Sargolzaei M, Plastow G, Wang Z, Karimi K, Hu G, Valipour S, Miar Y. Davoudi P, et al. BMC Genomics. 2022 Sep 13;23(1):649. doi: 10.1186/s12864-022-08874-1. BMC Genomics. 2022. PMID: 36096727 Free PMC article. - Human-specific CpG "beacons" identify loci associated with human-specific traits and disease.
Bell CG, Wilson GA, Butcher LM, Roos C, Walter L, Beck S. Bell CG, et al. Epigenetics. 2012 Oct;7(10):1188-99. doi: 10.4161/epi.22127. Epub 2012 Sep 11. Epigenetics. 2012. PMID: 22968434 Free PMC article. - Whole-genome detection of disease-associated deletions or excess homozygosity in a case-control study of rheumatoid arthritis.
Wu CC, Shete S, Jo EJ, Xu Y, Lu EY, Chen WV, Amos CI. Wu CC, et al. Hum Mol Genet. 2013 Mar 15;22(6):1249-61. doi: 10.1093/hmg/dds512. Epub 2012 Dec 6. Hum Mol Genet. 2013. PMID: 23223014 Free PMC article. - Accurate indel prediction using paired-end short reads.
Grimm D, Hagmann J, Koenig D, Weigel D, Borgwardt K. Grimm D, et al. BMC Genomics. 2013 Feb 27;14:132. doi: 10.1186/1471-2164-14-132. BMC Genomics. 2013. PMID: 23442375 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
- RC2 HG005552-01/HG/NHGRI NIH HHS/United States
- U01 HG005209-01/HG/NHGRI NIH HHS/United States
- P41 HG004221-03S2/HG/NHGRI NIH HHS/United States
- P41 HG004221-03S3/HG/NHGRI NIH HHS/United States
- P41 HG004221-02/HG/NHGRI NIH HHS/United States
- R01 GM081533/GM/NIGMS NIH HHS/United States
- P41 HG004221-03/HG/NHGRI NIH HHS/United States
- R01 GM081533-02/GM/NIGMS NIH HHS/United States
- 077009/Wellcome Trust/United Kingdom
- R01 GM081533-01A1/GM/NIGMS NIH HHS/United States
- 077192/Wellcome Trust/United Kingdom
- U54 HG003067/HG/NHGRI NIH HHS/United States
- R01 HG004719-01/HG/NHGRI NIH HHS/United States
- R01 HG004719/HG/NHGRI NIH HHS/United States
- R01 HG004719-04/HG/NHGRI NIH HHS/United States
- U01 HG005209-02/HG/NHGRI NIH HHS/United States
- RC2 HG005552/HG/NHGRI NIH HHS/United States
- R01 MH091350/MH/NIMH NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- R01 GM081533-04/GM/NIGMS NIH HHS/United States
- G1000758/MRC_/Medical Research Council/United Kingdom
- R01 GM059290/GM/NIGMS NIH HHS/United States
- R01 GM081533-03/GM/NIGMS NIH HHS/United States
- RC2 HG005552-02/HG/NHGRI NIH HHS/United States
- 077014/Wellcome Trust/United Kingdom
- R01 HG004719-02/HG/NHGRI NIH HHS/United States
- P41 HG004221-01/HG/NHGRI NIH HHS/United States
- 062023/Wellcome Trust/United Kingdom
- P41 HG004221-03S1/HG/NHGRI NIH HHS/United States
- 085532/Wellcome Trust/United Kingdom
- R01 HG004719-02S1/HG/NHGRI NIH HHS/United States
- R01 HG004719-03/HG/NHGRI NIH HHS/United States
- U01 HG005209/HG/NHGRI NIH HHS/United States
- R21 AA022707/AA/NIAAA NIH HHS/United States
- P41 HG004221/HG/NHGRI NIH HHS/United States
- G0701805/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials