Hepatitis C virus NS5A protein interacts with phosphatidylinositol 4-kinase type IIIalpha and regulates viral propagation - PubMed (original) (raw)
Hepatitis C virus NS5A protein interacts with phosphatidylinositol 4-kinase type IIIalpha and regulates viral propagation
Yun-Sook Lim et al. J Biol Chem. 2011.
Abstract
Hepatitis C Virus (HCV) nonstructural 5A (NS5A) is a pleiotropic protein involved in viral RNA replication and modulation of the cellular physiology in HCV-infected cells. To elucidate the mechanisms of the HCV life cycle, we identified cellular factors interacting with the NS5A protein in HCV-infected cells. Huh7.5 cells were electroporated with HCV Jc1 RNA. Cellular factors associated with HCV NS5A were identified by immunoprecipitation with Dynabead-conjugated NS5A antibody and LC-MS/MS. Phosphatidylinositol 4-kinase type IIIα (PI4KIIIα) was identified as a binding partner for the NS5A protein. NS5A derived from both genotypes 1b and 2a interacted with PI4KIIIα. NS5A interacted with PI4KIIIα through amino acids 401-600 of PI4KIIIα and domain I of NS5A. Interference of the protein interaction between NS5A and PI4KIIIα decreased HCV propagation. Knockdown of PI4KIIIα significantly reduced HCV replication in Huh7 cells harboring the subgenomic replicon and in Huh7.5 cells infected with cell culture grown virus (HCVcc). Silencing of PI4KIIIα further inhibited HCV release into the tissue culture medium. NS5A may recruit PI4KIIIα to the HCV RNA replication complex. These data suggest that PI4KIIIα is an essential host factor that supports HCV proliferation and therefore PI4KIIIα may be a legitimate target for anti-HCV therapy.
Figures
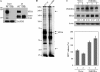
FIGURE 1.
Identification of the cellular binding partner for the HCV NS5A protein by immunoprecipitation. A, protein expression of NS5A and the specificity of NS5A antibody were confirmed by immunoprecipitation of Huh7.5 cells electroporated with either wild-type or replication defective Jc1 RNAs. GAPDH was used as a loading control. IP, immunoprecipitation; WT, wild-type; GND, replication defective mutant. B, Huh7.5 cells were transfected with either wild-type or replication defective Jc1 RNAs via electroporation. Cell lysates harvested at 4 days post-electroporation were immunoprecipitated with Dynabead-conjugated NS5A antibody. The immunoprecipitants were analyzed on 8% SDS-PAGE and silver stained. Interested bands were subjected to LC-MS/MS analysis. Asterisk denotes a nonspecific band. C, Huh7.5 cells stably expressing either p3XFLAG vector or p3XFLAG-PI4KIIIα were infected with HCVcc. Cells were harvested at day 2 postinfection and total cell lysates were immunoblotted with the indicated antibodies (top panel). HCV RNA release in vector stable and PI4KIIIα stable cells was determined by qPCR (bottom panel). *, p < 0.05, vector stable versus PI4KIIIα stable cells infected with HCVcc.
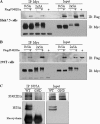
FIGURE 2.
NS5A interacts with PI4KIIIα. Either Huh7.5 (A) or HEK293T (B) cells were transfected with Myc-tagged NS5A of HCV genotypes 1b or 2a in the absence or presence of FLAG-tagged PI4KIIIα. Total cell lysates were immunoprecipitated (IP) with anti-Myc antibody, and bound proteins were immunoblotted (IB) with anti-FLAG antibody. Protein expressions of NS5A were confirmed using the same cell lysates by immunoblotting with anti-Myc antibody. C, Huh7.5 cells were infected with either wild-type or replication defective HCV and cell lysates harvested at 3 days after infection were immunoprecipitated with NS5A antibody. Bound proteins were immunoblotted with anti-PI4KIIIα antibody. Protein expressions of NS5A and PI4KIIIα were confirmed using the same cell lysates by immunoblotting with anti-NS5A and anti-PI4KIIIα antibody.
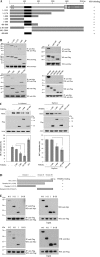
FIGURE 3.
NS5A interacts with PI4KIIIα through aa 401–600 of PI4KIIIα and domain I of NS5A. A, schematic diagram of both wild-type and mutant constructs of PI4KIIIα protein. LKU, lipid kinase unique domain; PH, pleckstrin homology; CAT, catalytic domain. B, HEK293T cells were cotransfected with Myc-tagged NS5A and FLAG-tagged mutant constructs of PI4KIIIα. Total cell lysates harvested at 24 h after transfection were immunoprecipitated (IP) with anti-Myc antibody and bound proteins were immunoblotted with anti-FLAG antibody (left, top panel). Protein expressions of both PI4KIIIα and NS5A were confirmed using the same cell lysates by immunoblotting with anti-FLAG and anti-Myc antibody, respectively (left, top panels). The NS5A binding region in PI4KIIIα was further defined by using more mutants (left, bottom, and right, top three panels) and then precisely determined using serially truncated mutants (right, bottom three panels). C, left panels, Huh7.5 cells were transfected with various truncated mutants of PI4KIIIα as indicated. At 24 h after transfection, cells were infected with HCVcc. Two days after HCV infection, cell lysates were immunoblotted with the indicated antibodies (top panels) and HCV RNA levels were determined by qPCR (bottom panel). *, p < 0.05, 1–400 versus 1–800 or 401–800. C, right panels, Huh7 cells harboring HCV replicon were transfected with various mutants of PI4KIIIα and harvested at 3 days after transfection. Cell lysates were immunoblotted with the indicated antibodies (top panels). Total RNAs extracted from either IFN-cured or HCV replicon cells were quantified by qRT-PCR (bottom panel), *, p < 0.05, 1–400 versus 401–800. D, schematic diagram shows both wild-type and mutant forms of the NS5A protein. E, Myc-tagged NS5A and FLAG-tagged PI4KIIIα proteins were coexpressed in HEK293T cells. PI4KIIIα was immunoprecipitated with anti-FLAG antibody and bound proteins were immunoblotted with anti-Myc antibody (left, top panel). Both protein expression and immunoprecipitation of PI4KIIIα were confirmed by anti-FLAG antibody. Protein expressions of both NS5A and PI4KIIIα were confirmed using the same cell lysates by immunoblotting with anti-Myc and anti-FLAG antibody, respectively (right, top panels). HEK293T cells were cotransfected with either Myc-tagged wild-type or mutants of NS5A and FLAG-tagged PI4KIIIα mutant (aa 400–600) expression plasmids. Coimmunoprecipitation and Western blot (WB) were performed as described above (bottom two panels).
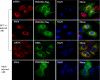
FIGURE 4.
PI4KIIIα is a component of the HCV RNA replication complex. Huh7.5 cells were either mock-infected or infected with HCVcc. At 2 days postinfection, cells were transfected with plasmid expressing FLAG-tagged PI4KIIIα. At 36 h after transfection, cells were fixed and incubated with the indicated antibodies for 2 h. After being washed with PBS, cells were further incubated with the appropriate secondary antibodies. Samples were analyzed for immunofluorescence staining using a Zeiss LSM 700 laser confocal microscopy system. Cells were counterstained with 4′6-diamidino-2-phenylindole (DAPI) to label nuclei.
FIGURE 5.
Silencing of PI4KIIIα reduces both protein expression and RNA replication levels in HCV subgenomic replicon cells. A, Huh7 cells harboring HCV subgenomic replicon were transfected with PI4KIIIα siRNA pool or the indicated control siRNA constructs. Total cell lysates harvested at 72 h after transfection were immunoblotted with the indicated antibodies. Negative, irrelevant siRNA; positive, HCV-specific siRNA. B, RNAs extracted from siRNA-transfected HCV replicon cells were measured by qRT-PCR at 96 h after transfection of PI4KIIIα siRNA. C, cDNAs were amplified by PCR and analyzed on an agarose gel.
FIGURE 6.
Knockdown of PI4KIIIα blocks HCV propagation. A, Huh7.5 cells were transfected with PI4KIIIα siRNAs or the indicated siRNA constructs and followed by HCV infection at 48 h after transfection. Total cell lysates harvested at 48 h after HCV infection were immunoblotted with the indicated antibodies. B, cell viability was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay at 96 h after transfection. C–E, Huh7.5 cells were transfected with the indicated siRNAs and infected with HCV at 48 h after transfection. HCV RNAs extracted from the cells (C) and cell culture medium (D) were measured by qRT-PCR. cDNA was synthesized by using RNA extracted from cell culture medium and amplified by PCR (E). F, Huh7.5 cells were transfected with the four individual siRNA of PI4KIIIα via electroporation, allowed silencing for 48 h, and then infected these cells with HCV. Total cell lysates harvested at 48 h after infection were immunoblotted with the indicated antibodies (top panel). PI4KIIIα mRNA levels in each sample were quantified by qPCR (bottom panel).
Similar articles
- The lipid kinase phosphatidylinositol-4 kinase III alpha regulates the phosphorylation status of hepatitis C virus NS5A.
Reiss S, Harak C, Romero-Brey I, Radujkovic D, Klein R, Ruggieri A, Rebhan I, Bartenschlager R, Lohmann V. Reiss S, et al. PLoS Pathog. 2013 May;9(5):e1003359. doi: 10.1371/journal.ppat.1003359. Epub 2013 May 9. PLoS Pathog. 2013. PMID: 23675303 Free PMC article. - Mapping of functional domains of the lipid kinase phosphatidylinositol 4-kinase type III alpha involved in enzymatic activity and hepatitis C virus replication.
Harak C, Radujkovic D, Taveneau C, Reiss S, Klein R, Bressanelli S, Lohmann V. Harak C, et al. J Virol. 2014 Sep 1;88(17):9909-26. doi: 10.1128/JVI.01063-14. Epub 2014 Jun 11. J Virol. 2014. PMID: 24920820 Free PMC article. - Phosphorylation of hepatitis C virus NS5A nonstructural protein: a new paradigm for phosphorylation-dependent viral RNA replication?
Huang Y, Staschke K, De Francesco R, Tan SL. Huang Y, et al. Virology. 2007 Jul 20;364(1):1-9. doi: 10.1016/j.virol.2007.01.042. Epub 2007 Apr 2. Virology. 2007. PMID: 17400273 Review. - Hepatitis C virus NS5A: tales of a promiscuous protein.
Macdonald A, Harris M. Macdonald A, et al. J Gen Virol. 2004 Sep;85(Pt 9):2485-2502. doi: 10.1099/vir.0.80204-0. J Gen Virol. 2004. PMID: 15302943 Review.
Cited by
- Coxsackievirus mutants that can bypass host factor PI4KIIIβ and the need for high levels of PI4P lipids for replication.
van der Schaar HM, van der Linden L, Lanke KH, Strating JR, Pürstinger G, de Vries E, de Haan CA, Neyts J, van Kuppeveld FJ. van der Schaar HM, et al. Cell Res. 2012 Nov;22(11):1576-92. doi: 10.1038/cr.2012.129. Epub 2012 Sep 4. Cell Res. 2012. PMID: 22945356 Free PMC article. - Saponin inhibits hepatitis C virus propagation by up-regulating suppressor of cytokine signaling 2.
Lee J, Lim S, Kang SM, Min S, Son K, Lee HS, Park EM, Ngo HT, Tran HT, Lim YS, Hwang SB. Lee J, et al. PLoS One. 2012;7(6):e39366. doi: 10.1371/journal.pone.0039366. Epub 2012 Jun 20. PLoS One. 2012. PMID: 22745742 Free PMC article. - The role of phosphatidylinositol 4-kinases and phosphatidylinositol 4-phosphate during viral replication.
Delang L, Paeshuyse J, Neyts J. Delang L, et al. Biochem Pharmacol. 2012 Dec 1;84(11):1400-8. doi: 10.1016/j.bcp.2012.07.034. Epub 2012 Aug 8. Biochem Pharmacol. 2012. PMID: 22885339 Free PMC article. Review. - Quantitative proteomic analysis of host-virus interactions reveals a role for Golgi brefeldin A resistance factor 1 (GBF1) in dengue infection.
Carpp LN, Rogers RS, Moritz RL, Aitchison JD. Carpp LN, et al. Mol Cell Proteomics. 2014 Nov;13(11):2836-54. doi: 10.1074/mcp.M114.038984. Epub 2014 May 22. Mol Cell Proteomics. 2014. PMID: 24855065 Free PMC article. - Mammalian phosphatidylinositol 4-kinases as modulators of membrane trafficking and lipid signaling networks.
Clayton EL, Minogue S, Waugh MG. Clayton EL, et al. Prog Lipid Res. 2013 Jul;52(3):294-304. doi: 10.1016/j.plipres.2013.04.002. Epub 2013 Apr 19. Prog Lipid Res. 2013. PMID: 23608234 Free PMC article. Review.
References
- Kim W. R. (2002) Hepatology 36, S30–S34 - PubMed
- El-Serag H. B. (2002) J. Clin. Gastroenterol. 35, S72–S78 - PubMed
- Shepard C. W., Finelli L., Alter M. J. (2005) Lancet Infect. Dis. 5, 558–567 - PubMed
- Park K. J., Choi S. H., Choi D. H., Park J. M., Yie S. W., Lee S. Y., Hwang S. B. (2003) J. Biol. Chem. 278, 30711–30718 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases