LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans - PubMed (original) (raw)
LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans
Priscilla M Van Wynsberghe et al. Nat Struct Mol Biol. 2011 Mar.
Abstract
The highly conserved let-7 microRNA (miRNA) regulates developmental pathways across animal phyla. Mis-expression of let-7 causes lethality in C. elegans and has been associated with several human diseases. We show that timing of let-7 expression in developing worms is under complex transcriptional and post-transcriptional control. Expression of let-7 primary transcripts oscillates during each larval stage, but precursor and mature let-7 miRNAs do not accumulate until later in development after LIN-28 protein has diminished. We demonstrate that LIN-28 binds endogenous primary let-7 transcripts co-transcriptionally. We further show that LIN-28 binds endogenous primary let-7 transcripts in the nuclear compartment of human ES cells, suggesting that this LIN-28 activity is conserved across species. We conclude that co-transcriptional interaction of LIN-28 with let-7 primary transcripts blocks Drosha processing and, thus, precocious expression of mature let-7 during early development.
Figures
Figure 1
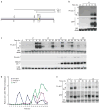
Expression of let-7 is transcriptionally and post-transcriptionally regulated. (a) Depiction of the 2460 nt long let-7 rescue construct with the positions of the mature let-7 sequence (shaded blue), 3′ splice site (SS; shaded yellow), two start sites (A and B) and approximate sizes of the spliced and unspliced transcripts indicated,. (b) Total RNA was isolated from embryos (E) or synchronized plet-7B∷GFP transgenic worms and analyzed by northern blotting. The similar sized B and SL1 transcripts often do not clearly resolve. (c) Total RNA was isolated from embryos (E) or synchronized WT N2 worms and analyzed by agarose or PAGE northern blotting. Representative blots from four independent experiments are shown. (d) Average pri-, pre- and mature let-7 levels after normalization to 18s or 5.8s rRNA from four independent experiments. (e) Total RNA was isolated from synchronized plet-7B∷GFP transgenic worms and analyzed as in Figure 1b. The entire blot is shown in Supplementary Figure 1c.
Figure 2
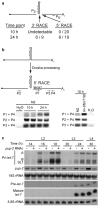
Developmentally regulated processing of let-7 pri-miRNA transcripts. (a) Depiction of expected Drosha cleavage products: 5′ flanking, let-7 hairpin precursor, and 3′ flanking. The number of sequenced RACE clones that mapped to the precise 3′ and 5′ Drosha cleavage products at each time point from two independent experiments is shown. The sequences of all Drosha cleavage products are shown in Supplementary Figure 3. (b) RT-PCR was performed on two independent 5′ RACE samples from N2 (left panel) or N2 and lin-28(n719) worms (right panel). (c) Total RNA was isolated from synchronized eri-1(mg366) RNAi hypersensitive worms at the indicated time points after vector control (-) or pup-2 (+) RNAi treatment, and analyzed by agarose and PAGE northern blotting. Representative blots from three independent experiments are shown.
Figure 3
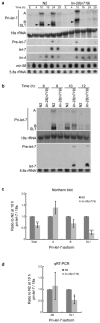
Regulation of let-7 processing by lin-28. (a) Total RNA was isolated from WT N2 or lin-28(n719) embryos (E) and synchronized worms at the indicated time points, and analyzed by agarose and PAGE northern blotting. Representative blots from three independent experiments are shown. The arrowheads mark the location of the SL1 pri-let-7 transcript. (b) Total RNA was isolated and analyzed as in Figure 3a. Representative blots from three independent experiments are shown. (c-d) Levels of each pri-let-7 isoform at the 10 h time point in lin-28(n719) relative to WT N2 worms after 18s rRNA normalization were calculated from six independent northern blot experiments (c) or 3 independent qRT-PCR experiments (d) and analyzed by Student's t-tests (*, p<0.05**, p<0.005; ***, p<0.0005). Error bars show s.e.m.
Figure 4
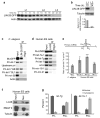
LIN-28 binds endogenous let-7 primary transcripts in C. elegans and human ES cells. (a-b) Total protein was isolated from PQ272 (LIN-28:GFP) worms and analyzed by western blotting. (b) Ratios of LIN-28:GFP levels to the 10 h time point after tubulin normalization were calculated from three independent experiments and analyzed by Student's t-tests (***, p<0.0005). Error bars show s.e.m. (c) Synchronized PQ272 worms were collected at 10 h and analyzed by RNA immunoprecipitation (RIP). Input, and LIN-28:GFP and IgG immunoprecipitates were analyzed by western blotting or RT-PCR. (d) Undifferentiated HUES6 cells were analyzed by RIP. Input, Lin28 and IgG immunoprecipitates were analyzed by western blotting or RT-PCR. (e) The worm and human cell samples from Figure 4c-d were analyzed by qRT-PCR to determine the levels of input or LIN-28 immunoprecipitated pri- or pre-let-7 RNAs using primers specific for pri-let-7 (priF and priR) or pre-let-7 and pri-let-7 transcripts containing the precursor hairpin (preF and preR). The ratio of precursor containing let-7 transcripts to pri-let-7 transcripts for immunoprecipitated samples after normalization to input samples for at least three independent experiments is shown, and was analyzed by Student's t-tests (*, p<0.05; **, p<0.005; ***, p<0.0005). Error bars show s.e.m. (f-g) Undifferentiated HUES6 cells were fractionationated into nuclear and cytoplasmic extracts before analysis by RIP and western blotting (f) or qRT-PCR as in Figure 4e (g). Results from three independent experiments are shown.
Figure 5
LIN-28 binds endogenous let-7 genomic DNA. Synchronized PQ272 (LIN-28:GFP) or pD4792 (GFP) worms were collected at 10 h and analyzed by chromatin immunoprecipitation (ChIP) with polyclonal antibodies against RNA pol II, GFP or IgG. The ratio of the indicated genomic DNA in the immunoprecipitated sample to the input sample for at least three independent experiments is shown and was analyzed by Student's t-tests (*, p<0.05). Error bars show s.e.m.
Similar articles
- Caenorhabditis elegans period homolog lin-42 regulates the timing of heterochronic miRNA expression.
McCulloch KA, Rougvie AE. McCulloch KA, et al. Proc Natl Acad Sci U S A. 2014 Oct 28;111(43):15450-5. doi: 10.1073/pnas.1414856111. Epub 2014 Oct 15. Proc Natl Acad Sci U S A. 2014. PMID: 25319259 Free PMC article. - acn-1, a C. elegans homologue of ACE, genetically interacts with the let-7 microRNA and other heterochronic genes.
Metheetrairut C, Ahuja Y, Slack FJ. Metheetrairut C, et al. Cell Cycle. 2017 Oct 2;16(19):1800-1809. doi: 10.1080/15384101.2017.1344798. Epub 2017 Sep 21. Cell Cycle. 2017. PMID: 28933985 Free PMC article. - The Doubletime Homolog KIN-20 Mainly Regulates let-7 Independently of Its Effects on the Period Homolog LIN-42 in Caenorhabditis elegans.
Rhodehouse K, Cascino K, Aseltine L, Padula A, Weinstein R, Spina JS, Olivero CE, Van Wynsberghe PM. Rhodehouse K, et al. G3 (Bethesda). 2018 Jul 31;8(8):2617-2629. doi: 10.1534/g3.118.200392. G3 (Bethesda). 2018. PMID: 29880558 Free PMC article. - Regulation of pre-miRNA processing.
Lehrbach NJ, Miska EA. Lehrbach NJ, et al. Adv Exp Med Biol. 2010;700:67-75. Adv Exp Med Biol. 2010. PMID: 21627031 Review. - The let-7 family of microRNAs.
Roush S, Slack FJ. Roush S, et al. Trends Cell Biol. 2008 Oct;18(10):505-16. doi: 10.1016/j.tcb.2008.07.007. Epub 2008 Sep 4. Trends Cell Biol. 2008. PMID: 18774294 Review.
Cited by
- The GATA factor elt-1 regulates C. elegans developmental timing by promoting expression of the let-7 family microRNAs.
Cohen ML, Kim S, Morita K, Kim SH, Han M. Cohen ML, et al. PLoS Genet. 2015 Mar 27;11(3):e1005099. doi: 10.1371/journal.pgen.1005099. eCollection 2015 Mar. PLoS Genet. 2015. PMID: 25816370 Free PMC article. - Lin28: primal regulator of growth and metabolism in stem cells.
Shyh-Chang N, Daley GQ. Shyh-Chang N, et al. Cell Stem Cell. 2013 Apr 4;12(4):395-406. doi: 10.1016/j.stem.2013.03.005. Cell Stem Cell. 2013. PMID: 23561442 Free PMC article. Review. - How does Lin28 let-7 control development and disease?
Thornton JE, Gregory RI. Thornton JE, et al. Trends Cell Biol. 2012 Sep;22(9):474-82. doi: 10.1016/j.tcb.2012.06.001. Epub 2012 Jul 9. Trends Cell Biol. 2012. PMID: 22784697 Free PMC article. Review. - hnRNP Q/SYNCRIP interacts with LIN28B and modulates the LIN28B/let-7 axis in human hepatoma cells.
Chang JJ, Lin T, Jhang XY, Chan SP. Chang JJ, et al. PLoS One. 2024 Jul 8;19(7):e0304947. doi: 10.1371/journal.pone.0304947. eCollection 2024. PLoS One. 2024. PMID: 38976670 Free PMC article. - Computational Tools for Stem Cell Biology.
Bian Q, Cahan P. Bian Q, et al. Trends Biotechnol. 2016 Dec;34(12):993-1009. doi: 10.1016/j.tibtech.2016.05.010. Epub 2016 Jun 15. Trends Biotechnol. 2016. PMID: 27318512 Free PMC article. Review.
References
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39. - PubMed
- Pasquinelli AE, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–9. - PubMed
- Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. - PubMed
- Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32GM087004/GM/NIGMS NIH HHS/United States
- GM071654/GM/NIGMS NIH HHS/United States
- F32 GM087004/GM/NIGMS NIH HHS/United States
- R01 GM071654-07/GM/NIGMS NIH HHS/United States
- T32 CA009523/CA/NCI NIH HHS/United States
- R01 GM071654/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials