Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage - PubMed (original) (raw)
Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage
Ruiying Wang et al. Structure. 2011.
Abstract
The CRISPRs (Clustered Regularly Interspaced Short Palindromic Repeats) found in prokaryotic genomes confer small RNA-mediated protection against viruses and other invaders. CRISPR loci contain iterations of a short repeat sequence alternating with small segments of varying invader-derived sequences. Distinct families of CRISPR-associated Cas proteins function to cleave within the repeat sequence of CRISPR transcripts and produce the individual invader-targeting crRNAs. Here, we report the crystal structure of Pyrococcus furiosus Cas6 bound with a repeat RNA at 3.2 Å resolution. In contrast to other Cas families of endonucleases, Cas6 clasps nucleotides 2-9 of the repeat RNA using its two ferredoxin-like domains, and the enzyme-anchored 5' end tethers the distal cleavage site of the RNA between nucleotides 22 and 23 to the predicted enzyme active site on the opposite side of the ferrodoxin-like domains. Our findings suggest a wrap-around mechanism for CRISPR RNA recognition and cleavage by Cas6 and related processing endonucleases.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
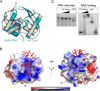
Fig. 1. Overview of CRISPR RNA processing steps and structural properties of Cas6
A, crRNA biogenesis. The primary CRISPR transcript contains repeats (R, red blocks) interspaced by guide sequences (G, grey blocks). The sequence of the repeat (RNA used in this study) is shown. Cas6 cleaves within each repeat (white dotted lines), releasing individual crRNA units (1X intermediates) that are further trimmed at the 3’ end by an unknown exonuclease(s). B, Overview of the Cas6-RNA complex structure shown in two orientations. The protein is shown in teal and RNA is in red. The α-helix α2 that contains the putative catalytic residue, His46, is indicated. Nucleotides of the repeat RNA are numbered from the 5’ end. See also Figures S1 and S2.
Fig. 2. Structural comparison of free and RNA-bound Cas6, the surface properties of the complex, and the effect of G-rich loop mutations
A, Superimposed structures of Cas6 bound to RNA (teal) and free Cas6 (grey). The putative catalytic residue (His46) is indicated in magenta. B, Electrostatic potentials of Cas6 in the RNA-bound complex are shown. The left view is the same as that in A. Cas6 binds nucleotides 2–10 of the repeat RNA (red) and is predicted to tether the 3’ end of the repeat RNA (dashed line) to its cleavage site (indicated by His46). Note that the potential path overlaps with the conserved Cas6-specific G-rich loop (G-loop). C, Impact on RNA cleavage (left) and binding (right) of Cas6 by alanine substation of Gly223, Gly225, Gly231, and Gly233 (G-loopm). The three concentrations of the G-loopm protein used in the cleavage and binding experiments were 0.5 µM, 1 µM and 10 µM, respectively.
Fig. 3. Key interaction features of the Cas6-RNA complex
The protein is represented by surface (upper panels) or stick models (lower panels) and the RNA is represented by red stick models. Contacts shorter than 3.4 Å between RNA and protein atoms are indicated by dashed lines. A, Interactions between Cas6 residues and nucleotides 2–6. B. Interactions between Cas6 residues and nucleotides 7–10.
Fig. 4. Detailed interactions between repeat RNA nucleotides and Cas6 residues
RNA nucleotides are represented by red sticks and protein residues are by grey-blue-red sticks. Nucleotides 4 and 9 involve simple interactions and are thus not included.
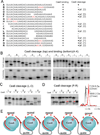
Fig. 5. Analysis of Cas6 binding and cleavage of a series of CRISPR repeat RNA mutants and a proposed model of Cas6 function
A, Sequences of the wild-type (A) and mutant repeat RNAs (B–R) used in Cas6 cleavage and binding studies. Mutant B (2’-deoxy modification on positions 22 and 23) is used as a non-cleavable substrate control as previously established by Carte et al. (Carte et al., 2008). Substituted, inserted, or modified nucleotides are shown in red. Deletions are indicated by spaces. “d” denotes 2’-deoxy modification. Results are summarized on the right. For mutations near the cleavage site, the experimentally observed cleavage position is denoted. B, Cleavage (upper panels) and binding (lower panels) of wild-type (A) and mutant RNAs (B–K) by Cas6. C, Cleavage and mass spectrometry analysis of the wild-type and mutant RNAs (L–O). D, Cleavage of repeat RNAs containing mutation A23U. Sizes of cleavage products are indicated. Analysis of the 21 and 22 nt cleavage products by MALDI-TOF mass spectrometric methods (Bruker, autoflex III) is shown in the right panel. The red trace is the product of cleavage of the wild-type RNA and the black trace is that from cleavage of the A23U mutant RNA. The predicted molecular weights of the 21 and 22 nt products, both with 2’,3’ cyclic phosphate termini, are indicated. E, Depiction of a beads-on-a-string model of CRISPR RNA processing by Cas6. CRISPR repeats are shown in red and invader-derived guide sequences are shown in grey. Individual Cas6 proteins bind the 5’ regions of the CRISPR repeats and the 3’ regions of the repeats wrap around Cas6 to the cleavage site (scissors). See also Figure S3.
Comment in
- Recognition of archaeal CRISPR RNA: No P in the alindromic repeat?
Lawrence CM, White MF. Lawrence CM, et al. Structure. 2011 Feb 9;19(2):142-4. doi: 10.1016/j.str.2011.01.003. Structure. 2011. PMID: 21300282
Similar articles
- Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes.
Carte J, Wang R, Li H, Terns RM, Terns MP. Carte J, et al. Genes Dev. 2008 Dec 15;22(24):3489-96. doi: 10.1101/gad.1742908. Genes Dev. 2008. PMID: 19141480 Free PMC article. - Evolution of CRISPR RNA recognition and processing by Cas6 endonucleases.
Niewoehner O, Jinek M, Doudna JA. Niewoehner O, et al. Nucleic Acids Res. 2014 Jan;42(2):1341-53. doi: 10.1093/nar/gkt922. Epub 2013 Oct 22. Nucleic Acids Res. 2014. PMID: 24150936 Free PMC article. - Substrate generation for endonucleases of CRISPR/cas systems.
Zoephel J, Dwarakanath S, Richter H, Plagens A, Randau L. Zoephel J, et al. J Vis Exp. 2012 Sep 8;(67):4277. doi: 10.3791/4277. J Vis Exp. 2012. PMID: 22986408 Free PMC article. - The RNA- and DNA-targeting CRISPR-Cas immune systems of Pyrococcus furiosus.
Terns RM, Terns MP. Terns RM, et al. Biochem Soc Trans. 2013 Dec;41(6):1416-21. doi: 10.1042/BST20130056. Biochem Soc Trans. 2013. PMID: 24256230 Free PMC article. Review. - CRISPR-based adaptive and heritable immunity in prokaryotes.
van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. van der Oost J, et al. Trends Biochem Sci. 2009 Aug;34(8):401-7. doi: 10.1016/j.tibs.2009.05.002. Epub 2009 Jul 29. Trends Biochem Sci. 2009. PMID: 19646880 Review.
Cited by
- Nature and intensity of selection pressure on CRISPR-associated genes.
Takeuchi N, Wolf YI, Makarova KS, Koonin EV. Takeuchi N, et al. J Bacteriol. 2012 Mar;194(5):1216-25. doi: 10.1128/JB.06521-11. Epub 2011 Dec 16. J Bacteriol. 2012. PMID: 22178975 Free PMC article. - The nuts and bolts of the Haloferax CRISPR-Cas system I-B.
Maier LK, Stachler AE, Brendel J, Stoll B, Fischer S, Haas KA, Schwarz TS, Alkhnbashi OS, Sharma K, Urlaub H, Backofen R, Gophna U, Marchfelder A. Maier LK, et al. RNA Biol. 2019 Apr;16(4):469-480. doi: 10.1080/15476286.2018.1460994. Epub 2018 May 21. RNA Biol. 2019. PMID: 29649958 Free PMC article. - Genome editing in mammalian cells using the CRISPR type I-D nuclease.
Osakabe K, Wada N, Murakami E, Miyashita N, Osakabe Y. Osakabe K, et al. Nucleic Acids Res. 2021 Jun 21;49(11):6347-6363. doi: 10.1093/nar/gkab348. Nucleic Acids Res. 2021. PMID: 34076237 Free PMC article. - A genome-wide view of the expression and processing patterns of Thermus thermophilus HB8 CRISPR RNAs.
Juranek S, Eban T, Altuvia Y, Brown M, Morozov P, Tuschl T, Margalit H. Juranek S, et al. RNA. 2012 Apr;18(4):783-94. doi: 10.1261/rna.031468.111. Epub 2012 Feb 21. RNA. 2012. PMID: 22355165 Free PMC article. - The impact of CRISPR repeat sequence on structures of a Cas6 protein-RNA complex.
Wang R, Zheng H, Preamplume G, Shao Y, Li H. Wang R, et al. Protein Sci. 2012 Mar;21(3):405-17. doi: 10.1002/pro.2028. Epub 2012 Feb 9. Protein Sci. 2012. PMID: 22238224 Free PMC article.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. - PubMed
- Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. - PubMed
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. - PubMed
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM054682-07/GM/NIGMS NIH HHS/United States
- R01 GM066958-07/GM/NIGMS NIH HHS/United States
- R01 GM054682/GM/NIGMS NIH HHS/United States
- R01 GM054682-13/GM/NIGMS NIH HHS/United States
- R01 GM099604/GM/NIGMS NIH HHS/United States
- R01 GM54682/GM/NIGMS NIH HHS/United States
- R01 GM054682-09S1/GM/NIGMS NIH HHS/United States
- R01 GM066958/GM/NIGMS NIH HHS/United States
- R01 GM054682-08/GM/NIGMS NIH HHS/United States
- R01 GM066958-07S1/GM/NIGMS NIH HHS/United States
- R01 GM054682-08S1/GM/NIGMS NIH HHS/United States
- R01 GM066958-08/GM/NIGMS NIH HHS/United States
- R01 GM054682-07S1/GM/NIGMS NIH HHS/United States
- R01 GM066958-06A1/GM/NIGMS NIH HHS/United States
- R01 GM066958-09/GM/NIGMS NIH HHS/United States
- R01 GM054682-06A1/GM/NIGMS NIH HHS/United States
- R01 GM054682-09/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources