Metabolic cross-talk allows labeling of O-linked beta-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway - PubMed (original) (raw)
Metabolic cross-talk allows labeling of O-linked beta-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway
Michael Boyce et al. Proc Natl Acad Sci U S A. 2011.
Abstract
Hundreds of mammalian nuclear and cytoplasmic proteins are reversibly glycosylated by O-linked β-N-acetylglucosamine (O-GlcNAc) to regulate their function, localization, and stability. Despite its broad functional significance, the dynamic and posttranslational nature of O-GlcNAc signaling makes it challenging to study using traditional molecular and cell biological techniques alone. Here, we report that metabolic cross-talk between the N-acetylgalactosamine salvage and O-GlcNAcylation pathways can be exploited for the tagging and identification of O-GlcNAcylated proteins. We found that N-azidoacetylgalactosamine (GalNAz) is converted by endogenous mammalian biosynthetic enzymes to UDP-GalNAz and then epimerized to UDP-N-azidoacetylglucosamine (GlcNAz). O-GlcNAc transferase accepts UDP-GlcNAz as a nucleotide-sugar donor, appending an azidosugar onto its native substrates, which can then be detected by covalent labeling using azide-reactive chemical probes. In a proof-of-principle proteomics experiment, we used metabolic GalNAz labeling of human cells and a bioorthogonal chemical probe to affinity-purify and identify numerous O-GlcNAcylated proteins. Our work provides a blueprint for a wide variety of future chemical approaches to identify, visualize, and characterize dynamic O-GlcNAc signaling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
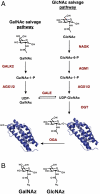
(A) The GlcNAc and GalNAc salvage and O-GlcNAc signaling pathways. Enzyme names shown in red. (B) GalNAz and GlcNAz.
Fig. 2.
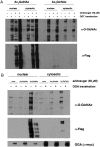
Metabolic labeling by GalNAz, but not GlcNAz, robustly mimics natural O-GlcNAc. 293T cells were mock-transfected or transfected with a construct expressing (A) OGT or (B) OGA and treated with vehicle or azidosugar for 24 h. Nuclear and cytoplasmic extracts were prepared, reacted with phosphine-Flag, and analyzed by immunoblot.
Fig. 3.
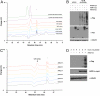
GlcNAz does not transit the UDP-GlcNAc pyrophosphorylase step in the GlcNAc salvage pathway. (A) Jurkat or 293T cells were treated with vehicle or 100 μM azidosugar for 24 h. Ethanol extracts were made and analyzed by HPAEC. Synthetic UDP-GlcNAz and UDP-GalNAz standards were included. Asterisk: an unknown species present in all cell-derived samples. (B) 293T cells were mock-transfected or transfected with the expression construct indicated and treated with vehicle or Ac4GlcNAz for 24 h. Cell lysates were reacted with phosphine-Flag and analyzed by immunoblot. (C) 293T cells were mock-transfected or transfected with an AGX2-myc expression construct. After 21 h, all cells were treated with 100 μM Ac4GlcNAz, samples were harvested by ethanol extraction after 3, 6, or 9 additional hours and analyzed by HPAEC. Asterisk: an unknown species present in all cell-derived samples. (D) Ethanol-insoluble protein fractions from the samples in C were resuspended in 8 M urea, reacted with phosphine-Flag, and analyzed by immunoblot.
Fig. 4.
Ac4GalNAz treatment results in GALE-dependent UDP-GlcNAz biosynthesis and labeling of O-GlcNAcylated proteins. (A) CHO or ldlD CHO cells were treated with vehicle or Ac4GalNAz for 24 h. Nuclear and cytoplasmic extracts were prepared, reacted with phosphine-Flag, and analyzed by immunoblot. Total protein was visualized by India ink staining. (B) CHO or ldlD mutant CHO cells were treated with vehicle or Ac4GalNAz for 72 h. Cells were fixed and azidoglycans detected via reaction with phosphine-Flag and immunofluorescence microscopy using an anti-Flag-FITC antibody conjugate. A Hoechst 33258 stain was included to visualize nuclei. (Top) FITC only. (Bottom): FITC/Hoechst merge.
Similar articles
- [Comparison of the performance of secretome analysis based on metabolic labeling by three unnatural sugars].
Mao Y, Zheng J, Feng S, Tian R. Mao Y, et al. Se Pu. 2021 Oct;39(10):1086-1093. doi: 10.3724/SP.J.1123.2021.04017. Se Pu. 2021. PMID: 34505430 Free PMC article. Chinese. - [Precise identification of _O_-linked _β_-_N_-acetylglucosamine peptides based on _O_-mesitylenesulfonylhydroxylamine elimination reaction].
Guo Z, Li H, Qin W. Guo Z, et al. Se Pu. 2021 Nov;39(11):1182-1190. doi: 10.3724/SP.J.1123.2020.12024. Se Pu. 2021. PMID: 34677013 Free PMC article. Chinese. - Identification of O-linked N-acetylglucosamine (O-GlcNAc)-modified osteoblast proteins by electron transfer dissociation tandem mass spectrometry reveals proteins critical for bone formation.
Nagel AK, Schilling M, Comte-Walters S, Berkaw MN, Ball LE. Nagel AK, et al. Mol Cell Proteomics. 2013 Apr;12(4):945-55. doi: 10.1074/mcp.M112.026633. Epub 2013 Feb 26. Mol Cell Proteomics. 2013. PMID: 23443134 Free PMC article. - Chemical Reporters and Their Bioorthogonal Reactions for Labeling Protein _O_-GlcNAcylation.
Kim EJ. Kim EJ. Molecules. 2018 Sep 20;23(10):2411. doi: 10.3390/molecules23102411. Molecules. 2018. PMID: 30241321 Free PMC article. Review. - Intracellular protein O-GlcNAc modification integrates nutrient status with transcriptional and metabolic regulation.
Nagel AK, Ball LE. Nagel AK, et al. Adv Cancer Res. 2015;126:137-66. doi: 10.1016/bs.acr.2014.12.003. Epub 2015 Feb 7. Adv Cancer Res. 2015. PMID: 25727147 Review.
Cited by
- Chemistry-enabled methods for the visualization of cell-surface glycoproteins in Metazoans.
Chuh KN, Pratt MR. Chuh KN, et al. Glycoconj J. 2015 Oct;32(7):443-54. doi: 10.1007/s10719-015-9589-3. Epub 2015 Apr 28. Glycoconj J. 2015. PMID: 25913724 Review. - In vivo metabolic labeling of sialoglycans in the mouse brain by using a liposome-assisted bioorthogonal reporter strategy.
Xie R, Dong L, Du Y, Zhu Y, Hua R, Zhang C, Chen X. Xie R, et al. Proc Natl Acad Sci U S A. 2016 May 10;113(19):5173-8. doi: 10.1073/pnas.1516524113. Epub 2016 Apr 28. Proc Natl Acad Sci U S A. 2016. PMID: 27125855 Free PMC article. - Methods for Enrichment and Assignment of N-Acetylglucosamine Modification Sites.
Maynard JC, Chalkley RJ. Maynard JC, et al. Mol Cell Proteomics. 2021;20:100031. doi: 10.1074/mcp.R120.002206. Epub 2021 Feb 9. Mol Cell Proteomics. 2021. PMID: 32938750 Free PMC article. Review. - Metabolic labeling of Caenorhabditis elegans primary embryonic cells with azido-sugars as a tool for glycoprotein discovery.
Burnham-Marusich AR, Snodgrass CJ, Johnson AM, Kiyoshi CM, Buzby SE, Gruner MR, Berninsone PM. Burnham-Marusich AR, et al. PLoS One. 2012;7(11):e49020. doi: 10.1371/journal.pone.0049020. Epub 2012 Nov 12. PLoS One. 2012. PMID: 23152843 Free PMC article. - Realizing the Promise of Chemical Glycobiology.
Wang LX, Davis BG. Wang LX, et al. Chem Sci. 2013 Sep 1;4(9):3381-3394. doi: 10.1039/C3SC50877C. Chem Sci. 2013. PMID: 23914294 Free PMC article.
References
- Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. - PubMed
- Yuzwa SA, et al. A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat Chem Biol. 2008;4:483–490. - PubMed
- Kneass ZT, Marchase RB. Neutrophils exhibit rapid agonist-induced increases in protein-associated O-GlcNAc. J Biol Chem. 2004;279:45759–45765. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM066047/GM/NIGMS NIH HHS/United States
- GM069157/GM/NIGMS NIH HHS/United States
- F32 GM069157/GM/NIGMS NIH HHS/United States
- PC080659/PC/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 GM066047/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources