Complete genome sequence of Brachybacterium faecium type strain (Schefferle 6-10) - PubMed (original) (raw)
Rüdiger Pukall, Kurt Labuttii, Alex Copeland, Tijana Glavina Del Rio, Matt Nolan, Feng Chen, Susan Lucas, Hope Tice, Jan-Fang Cheng, David Bruce, Lynne Goodwin, Sam Pitluck, Manfred Rohde, Markus Göker, Amrita Pati, Natalia Ivanova, Konstantinos Mavrommatis, Amy Chen, Krishna Palaniappan, Patrik D'haeseleer, Patrick Chain, Jim Bristow, Jonathan A Eisen, Victor Markowitz, Philip Hugenholtz, Nikos C Kyrpides, Hans-Peter Klenk
- PMID: 21304631
- PMCID: PMC3035206
- DOI: 10.4056/sigs.492
Complete genome sequence of Brachybacterium faecium type strain (Schefferle 6-10)
Alla Lapidus et al. Stand Genomic Sci. 2009.
Abstract
Brachybacterium faecium Collins et al. 1988 is the type species of the genus, and is of phylogenetic interest because of its location in the Dermabacteraceae, a rather isolated family within the actinobacterial suborder Micrococcineae. B. faecium is known for its rod-coccus growth cycle and the ability to degrade uric acid. It grows aerobically or weakly anaerobically. The strain described in this report is a free-living, nonmotile, Gram-positive bacterium, originally isolated from poultry deep litter. Here we describe the features of this organism, together with the complete genome sequence, and annotation. This is the first complete genome sequence of a member of the actinobacterial family Dermabacteraceae, and the 3,614,992 bp long single replicon genome with its 3129 protein-coding and 69 RNA genes is part of the Genomic Encyclopedia of Bacteria and Archaea project.
Keywords: Dermabacteraceae; aerobic; free-living; mesophile; non-pathogenic; rod-coccus growth cycle; uric acid degradation.
Figures
Figure 1
Phylogenetic tree of B. faecium Schefferle 6-10T and all type strains of the genus Brachybacterium, inferred from 1408 aligned characters [2] of the 16S rRNA sequence under the maximum likelihood criterion [3,4]. The tree was rooted with Dermabacter hominis, another member of the family Dermabacteraceae. The branches are scaled in terms of the expected number of substitutions per site. Numbers above branches are support values from 1000 bootstrap replicates, if larger than 60%. Strains with a genome-sequencing project registered in GOLD [5] are printed in blue; published genomes in bold.
Figure 2
Scanning electron micrograph of B. faecium Schefferle 6-10T
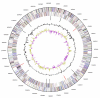
Figure 3
Graphical circular map of the genome. From outside to the center: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Figure 4
Schematic cellular overview of all pathways of the B. faecium strain Schefferle 6-10T metabolism. Nodes represent metabolites, with shape indicating class of metabolite. Lines represent reactions.
Similar articles
- Complete genome sequence of Beutenbergia cavernae type strain (HKI 0122).
Land M, Pukall R, Abt B, Göker M, Rohde M, Glavina Del Rio T, Tice H, Copeland A, Cheng JF, Lucas S, Chen F, Nolan M, Bruce D, Goodwin L, Pitluck S, Ivanova N, Mavromatis K, Ovchinnikova G, Pati A, Chen A, Palaniappan K, Hauser L, Chang YJ, Jefferies CC, Saunders E, Brettin T, Detter JC, Han C, Chain P, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP, Lapidus A. Land M, et al. Stand Genomic Sci. 2009 Jul 20;1(1):21-8. doi: 10.4056/sigs.1162. Stand Genomic Sci. 2009. PMID: 21304633 Free PMC article. - Complete genome sequence of Kytococcus sedentarius type strain (541).
Sims D, Brettin T, Detter JC, Han C, Lapidus A, Copeland A, Glavina Del Rio T, Nolan M, Chen F, Lucas S, Tice H, Cheng JF, Bruce D, Goodwin L, Pitluck S, Ovchinnikova G, Pati A, Ivanova N, Mavrommatis K, Chen A, Palaniappan K, D'haeseleer P, Chain P, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Schneider S, Göker M, Pukall R, Kyrpides NC, Klenk HP. Sims D, et al. Stand Genomic Sci. 2009 Jul 20;1(1):12-20. doi: 10.4056/sigs.761. Stand Genomic Sci. 2009. PMID: 21304632 Free PMC article. - Complete genome sequence of Actinosynnema mirum type strain (101).
Land M, Lapidus A, Mayilraj S, Chen F, Copeland A, Del Rio TG, Nolan M, Lucas S, Tice H, Cheng JF, Chertkov O, Bruce D, Goodwin L, Pitluck S, Rohde M, Göker M, Pati A, Ivanova N, Mavromatis K, Chen A, Palaniappan K, Hauser L, Chang YJ, Jeffries CC, Brettin T, Detter JC, Han C, Chain P, Tindall BJ, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP. Land M, et al. Stand Genomic Sci. 2009 Jul 20;1(1):46-53. doi: 10.4056/sigs.21137. Stand Genomic Sci. 2009. PMID: 21304636 Free PMC article. - Complete genome sequence of Halomicrobium mukohataei type strain (arg-2).
Tindall BJ, Schneider S, Lapidus A, Copeland A, Glavina Del Rio T, Nolan M, Lucas S, Chen F, Tice H, Cheng JF, Saunders E, Bruce D, Goodwin L, Pitluck S, Mikhailova N, Pati A, Ivanova N, Mavrommatis K, Chen A, Palaniappan K, Chain P, Land M, Hauser L, Chang YJ, Jeffries CD, Brettin T, Han C, Rohde M, Göker M, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Klenk HP, Kyrpides NC, Detter JC. Tindall BJ, et al. Stand Genomic Sci. 2009 Nov 22;1(3):270-7. doi: 10.4056/sigs.42644. Stand Genomic Sci. 2009. PMID: 21304667 Free PMC article. - Complete genome sequence of Catenulispora acidiphila type strain (ID 139908).
Copeland A, Lapidus A, Glavina Del Rio T, Nolan M, Lucas S, Chen F, Tice H, Cheng JF, Bruce D, Goodwin L, Pitluck S, Mikhailova N, Pati A, Ivanova N, Mavromatis K, Chen A, Palaniappan K, Chain P, Land M, Hauser L, Chang YJ, Jeffries CD, Chertkov O, Brettin T, Detter JC, Han C, Ali Z, Tindall BJ, Göker M, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP. Copeland A, et al. Stand Genomic Sci. 2009 Sep 24;1(2):119-25. doi: 10.4056/sigs.17259. Stand Genomic Sci. 2009. PMID: 21304647 Free PMC article.
Cited by
- ExSPAnder: a universal repeat resolver for DNA fragment assembly.
Prjibelski AD, Vasilinetc I, Bankevich A, Gurevich A, Krivosheeva T, Nurk S, Pham S, Korobeynikov A, Lapidus A, Pevzner PA. Prjibelski AD, et al. Bioinformatics. 2014 Jun 15;30(12):i293-301. doi: 10.1093/bioinformatics/btu266. Bioinformatics. 2014. PMID: 24931996 Free PMC article. - Genome sequence of Brachybacterium squillarum M-6-3(T), isolated from salt-fermented seafood.
Park SK, Roh SW, Whon TW, Bae JW. Park SK, et al. J Bacteriol. 2011 Nov;193(22):6416-7. doi: 10.1128/JB.06183-11. J Bacteriol. 2011. PMID: 22038973 Free PMC article. - Draft Genome Sequence of Brachybacterium phenoliresistens Strain W13A50, a Halotolerant Hydrocarbon-Degrading Bacterium.
Wang X, Zhang Z, Jin D, Zhou L, Wu L, Li C, Zhao L, An W, Chen Y. Wang X, et al. Genome Announc. 2014 Sep 18;2(5):e00899-14. doi: 10.1128/genomeA.00899-14. Genome Announc. 2014. PMID: 25237019 Free PMC article. - Fast Mechanically Driven Daughter Cell Separation Is Widespread in Actinobacteria.
Zhou X, Halladin DK, Theriot JA. Zhou X, et al. mBio. 2016 Aug 30;7(4):e00952-16. doi: 10.1128/mBio.00952-16. mBio. 2016. PMID: 27578753 Free PMC article. - The state of standards in genomic sciences.
Garrity GM. Garrity GM. Stand Genomic Sci. 2011 Dec 31;5(3):262-8. doi: 10.4650/sigs.2515706. Stand Genomic Sci. 2011. PMID: 22675577 Free PMC article. No abstract available.
References
- Collins M, Brown J, Jones D. Brachybacterium faecium gen. nov., sp. nov., a coryneform bacterium from poultry deep litter. Int J Syst Bacteriol 1988; 38:45-48
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases