Complete genome sequence of Olsenella uli type strain (VPI D76D-27C) - PubMed (original) (raw)
. 2010 Aug 20;3(1):76-84.
doi: 10.4056/sigs.1082860.
Brittany Held, Susan Lucas, Matt Nolan, Montri Yasawong, Tijana Glavina Del Rio, Hope Tice, Jan-Fang Cheng, David Bruce, John C Detter, Roxanne Tapia, Cliff Han, Lynne Goodwin, Sam Pitluck, Konstantinos Liolios, Natalia Ivanova, Konstantinos Mavromatis, Natalia Mikhailova, Amrita Pati, Amy Chen, Krishna Palaniappan, Miriam Land, Loren Hauser, Yun-Juan Chang, Cynthia D Jeffries, Manfred Rohde, Johannes Sikorski, Rüdiger Pukall, Tanja Woyke, James Bristow, Jonathan A Eisen, Victor Markowitz, Philip Hugenholtz, Nikos C Kyrpides, Hans-Peter Klenk, Alla Lapidus
- PMID: 21304694
- PMCID: PMC3035265
- DOI: 10.4056/sigs.1082860
Complete genome sequence of Olsenella uli type strain (VPI D76D-27C)
Markus Göker et al. Stand Genomic Sci. 2010.
Abstract
Olsenella uli (Olsen et al. 1991) Dewhirst et al. 2001 is the type species of the genus Olsenella, which belongs to the actinobacterial family Coriobacteriaceae. The species is of interest because it is frequently isolated from dental plaque in periodontitis patients and can cause primary endodontic infection. The species is a Gram-positive, non-motile and non-sporulating bacterium. The strain described in this study was isolated from human gingival crevices. This is the first completed sequence of the genus Olsenella and the fifth sequence from a member of the family Coriobacteriaceae. The 2,051,896 bp long genome with its 1,795 protein-coding and 55 RNA genes is a part of the Genomic Encyclopedia of Bacteria and Archaea project.
Keywords: Coriobacteriaceae; GEBA; human gingival crevices; microaerotolerant anaerobe; primary endodontic infections.
Figures
Figure 1
Scanning electron micrograph of O. uli VPI D76D-27CT
Figure 2
Phylogenetic tree highlighting the position of O. uli VPI D76D-27CT relative to the type strains within the genus and the type strains of the other genera within the family Coriobacteriaceae. The trees were inferred from 1,408 aligned characters [24,25] of the 16S rRNA gene sequence under the maximum likelihood criterion [26] and as far as possible (note: Olsenella is paraphyletic in this tree) rooted in accordance with the current taxonomy [27]. The branches are scaled in terms of the expected number of substitutions per site. Numbers above branches are support values from 250 bootstrap replicates [28] if larger than 60%. Lineages with type strain genome sequencing projects registered in GOLD [29] are shown in blue, published genomes in bold [30,31]. Adding the 16S rRNA sequence of the type strain of the not yet validly published species ‘Olsenella umbonata’ (FN178463) to the tree (data not shown) did not change the overall arrangement; ‘O. umbonata’ appeared within the grade between O. uli and O. profusa.
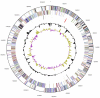
Figure 3
Graphical circular map of the genome. From outside to the center: Genes on forward strand (color by COG categories), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, rRNAs red, other RNAs black), GC content, GC skew.
Similar articles
- Complete genome sequence of Eggerthella lenta type strain (IPP VPI 0255).
Saunders E, Pukall R, Abt B, Lapidus A, Glavina Del Rio T, Copeland A, Tice H, Cheng JF, Lucas S, Chen F, Nolan M, Bruce D, Goodwin L, Pitluck S, Ivanova N, Mavromatis K, Ovchinnikova G, Pati A, Chen A, Palaniappan K, Land M, Hauser L, Chang YJ, Jeffries CD, Chain P, Meincke L, Sims D, Brettin T, Detter JC, Göker M, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP, Han C. Saunders E, et al. Stand Genomic Sci. 2009 Sep 28;1(2):174-82. doi: 10.4056/sigs.33592. Stand Genomic Sci. 2009. PMID: 21304654 Free PMC article. - Olsenella umbonata sp. nov., a microaerotolerant anaerobic lactic acid bacterium from the sheep rumen and pig jejunum, and emended descriptions of Olsenella, Olsenella uli and Olsenella profusa.
Kraatz M, Wallace RJ, Svensson L. Kraatz M, et al. Int J Syst Evol Microbiol. 2011 Apr;61(Pt 4):795-803. doi: 10.1099/ijs.0.022954-0. Epub 2010 Apr 30. Int J Syst Evol Microbiol. 2011. PMID: 20435744 - Complete genome sequence of Atopobium parvulum type strain (IPP 1246).
Copeland A, Sikorski J, Lapidus A, Nolan M, Del Rio TG, Lucas S, Chen F, Tice H, Pitluck S, Cheng JF, Pukall R, Chertkov O, Brettin T, Han C, Detter JC, Kuske C, Bruce D, Goodwin L, Ivanova N, Mavromatis K, Mikhailova N, Chen A, Palaniappan K, Chain P, Rohde M, Göker M, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP, Detter JC. Copeland A, et al. Stand Genomic Sci. 2009 Sep 23;1(2):166-73. doi: 10.4056/sigs.29547. Stand Genomic Sci. 2009. PMID: 21304653 Free PMC article. - Complete genome sequence of Arcanobacterium haemolyticum type strain (11018).
Yasawong M, Teshima H, Lapidus A, Nolan M, Lucas S, Glavina Del Rio T, Tice H, Cheng JF, Bruce D, Detter C, Tapia R, Han C, Goodwin L, Pitluck S, Liolios K, Ivanova N, Mavromatis K, Mikhailova N, Pati A, Chen A, Palaniappan K, Land M, Hauser L, Chang YJ, Jeffries CD, Rohde M, Sikorski J, Pukall R, Göker M, Woyke T, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP. Yasawong M, et al. Stand Genomic Sci. 2010 Sep 28;3(2):126-35. doi: 10.4056/sigs.1123072. Stand Genomic Sci. 2010. PMID: 21304742 Free PMC article.
Cited by
- Changes in the rumen development, rumen fermentation, and rumen microbiota community in weaned calves during steviol glycosides treatment.
Wang K, Jiang M, Chen Y, Huang Y, Cheng Z, Datsomor O, Jama SM, Zhu L, Li Y, Zhao G, Lin M. Wang K, et al. Front Microbiol. 2024 Jun 24;15:1395665. doi: 10.3389/fmicb.2024.1395665. eCollection 2024. Front Microbiol. 2024. PMID: 38979539 Free PMC article. - Enhancing the Conventional Culture: the Evaluation of Several Culture Media and Growth Conditions Improves the Isolation of Ruminal Bacteria.
Botero Rute LM, Caro-Quintero A, Acosta-González A. Botero Rute LM, et al. Microb Ecol. 2023 Dec 12;87(1):13. doi: 10.1007/s00248-023-02319-2. Microb Ecol. 2023. PMID: 38082143 Free PMC article. - Effect of grape pomace supplement on growth performance, gastrointestinal microbiota, and methane production in Tan lambs.
Cheng X, Du X, Liang Y, Degen AA, Wu X, Ji K, Gao Q, Xin G, Cong H, Yang G. Cheng X, et al. Front Microbiol. 2023 Sep 28;14:1264840. doi: 10.3389/fmicb.2023.1264840. eCollection 2023. Front Microbiol. 2023. PMID: 37840727 Free PMC article. - Influence of Parturition on Rumen Bacteria and SCFAs in Holstein Cows Based on 16S rRNA Sequencing and Targeted Metabolomics.
Guo Y, Wang F, Mao Y, Kong W, Wang J, Zhang G. Guo Y, et al. Animals (Basel). 2023 Feb 21;13(5):782. doi: 10.3390/ani13050782. Animals (Basel). 2023. PMID: 36899639 Free PMC article. - Analysis of serum antioxidant capacity and gut microbiota in calves at different growth stages in Tibet.
Zhang X, Cao Z, Yang H, Wang Y, Wang W, Li S. Zhang X, et al. Front Microbiol. 2023 Jan 30;13:1089488. doi: 10.3389/fmicb.2022.1089488. eCollection 2022. Front Microbiol. 2023. PMID: 36798869 Free PMC article.
References
- Stackebrandt E, Rainey FA, Ward-Rainey NL. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol 1997; 47:479-491 10.1099/00207713-47-2-479 - DOI
- Zhi XY, Li WJ, Stackebrandt E. An update of the structure and 16S rRNA gene sequence-based definition of higher ranks of the class Actinobacteria, with the proposal of two new suborders and four new families and emended descriptions of the existing higher taxa. Int J Syst Evol Microbiol 2009; 59:589-608 10.1099/ijs.0.65780-0 - DOI - PubMed
- Dewhirst FE, Paster BJ, Tzellas N, Coleman B, Downes J, Spratt DA, Wade WG. Characterization of novel human oral isolates and cloned 16S rDNA sequences that fall in the family Coriobacteriaceae: description of Olsenella gen. nov., reclassification of Lactobacillus uli as Olsenella uli comb. nov. and description of Olsenella profusa sp. nov. Int J Syst Evol Microbiol 2001; 51:1797-1804 - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases