Identification of candidate small-molecule therapeutics to cancer by gene-signature perturbation in connectivity mapping - PubMed (original) (raw)
Identification of candidate small-molecule therapeutics to cancer by gene-signature perturbation in connectivity mapping
Darragh G McArt et al. PLoS One. 2011.
Abstract
Connectivity mapping is a recently developed technique for discovering the underlying connections between different biological states based on gene-expression similarities. The sscMap method has been shown to provide enhanced sensitivity in mapping meaningful connections leading to testable biological hypotheses and in identifying drug candidates with particular pharmacological and/or toxicological properties. Challenges remain, however, as to how to prioritise the large number of discovered connections in an unbiased manner such that the success rate of any following-up investigation can be maximised. We introduce a new concept, gene-signature perturbation, which aims to test whether an identified connection is stable enough against systematic minor changes (perturbation) to the gene-signature. We applied the perturbation method to three independent datasets obtained from the GEO database: acute myeloid leukemia (AML), cervical cancer, and breast cancer treated with letrozole. We demonstrate that the perturbation approach helps to identify meaningful biological connections which suggest the most relevant candidate drugs. In the case of AML, we found that the prevalent compounds were retinoic acids and PPAR activators. For cervical cancer, our results suggested that potential drugs are likely to involve the EGFR pathway; and with the breast cancer dataset, we identified candidates that are involved in prostaglandin inhibition. Thus the gene-signature perturbation approach added real values to the whole connectivity mapping process, allowing for increased specificity in the identification of possible therapeutic candidates.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
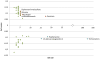
Figure 1. setscore-vs-setsize plot of significant connections to the cervical cancer signature.
Green- significant connections; Blue- significant negative connection setscores with high PS and large setszie; Red- significant positive connection setscores with high PS and large setsize.
Figure 2. Significant connections to the letrozole treatment signature in breast cancer.
Green- significant connections; Blue- significant positive connection setscores with high PS and large setsize; Red- significant negative connection setscores with high PS and large setsize.
Similar articles
- A gene-signature progression approach to identifying candidate small-molecule cancer therapeutics with connectivity mapping.
Wen Q, Kim CS, Hamilton PW, Zhang SD. Wen Q, et al. BMC Bioinformatics. 2016 May 11;17(1):211. doi: 10.1186/s12859-016-1066-x. BMC Bioinformatics. 2016. PMID: 27170106 Free PMC article. - Connectivity mapping using a combined gene signature from multiple colorectal cancer datasets identified candidate drugs including existing chemotherapies.
Wen Q, O'Reilly P, Dunne PD, Lawler M, Van Schaeybroeck S, Salto-Tellez M, Hamilton P, Zhang SD. Wen Q, et al. BMC Syst Biol. 2015;9 Suppl 5(Suppl 5):S4. doi: 10.1186/1752-0509-9-S5-S4. Epub 2015 Sep 1. BMC Syst Biol. 2015. PMID: 26356760 Free PMC article. - An integrated meta-analysis approach to identifying medications with potential to alter breast cancer risk through connectivity mapping.
Thillaiyampalam G, Liberante F, Murray L, Cardwell C, Mills K, Zhang SD. Thillaiyampalam G, et al. BMC Bioinformatics. 2017 Dec 21;18(1):581. doi: 10.1186/s12859-017-1989-x. BMC Bioinformatics. 2017. PMID: 29268695 Free PMC article. - Therapeutic Target Discovery Using High-Throughput Genetic Screens in Acute Myeloid Leukemia.
Liu Q, Garcia M, Wang S, Chen CW. Liu Q, et al. Cells. 2020 Aug 12;9(8):1888. doi: 10.3390/cells9081888. Cells. 2020. PMID: 32806592 Free PMC article. Review. - Network pharmacology of cancer: From understanding of complex interactomes to the design of multi-target specific therapeutics from nature.
Poornima P, Kumar JD, Zhao Q, Blunder M, Efferth T. Poornima P, et al. Pharmacol Res. 2016 Sep;111:290-302. doi: 10.1016/j.phrs.2016.06.018. Epub 2016 Jun 18. Pharmacol Res. 2016. PMID: 27329331 Review.
Cited by
- A gene-signature progression approach to identifying candidate small-molecule cancer therapeutics with connectivity mapping.
Wen Q, Kim CS, Hamilton PW, Zhang SD. Wen Q, et al. BMC Bioinformatics. 2016 May 11;17(1):211. doi: 10.1186/s12859-016-1066-x. BMC Bioinformatics. 2016. PMID: 27170106 Free PMC article. - Cystic Fibrosis from Laboratory to Bedside: The Role of A20 in NF-κB-Mediated Inflammation.
Bannon A, Zhang SD, Schock BC, Ennis M. Bannon A, et al. Med Princ Pract. 2015;24(4):301-10. doi: 10.1159/000381423. Epub 2015 Apr 25. Med Princ Pract. 2015. PMID: 25925366 Free PMC article. Review. - CLC-Pred: A freely available web-service for in silico prediction of human cell line cytotoxicity for drug-like compounds.
Lagunin AA, Dubovskaja VI, Rudik AV, Pogodin PV, Druzhilovskiy DS, Gloriozova TA, Filimonov DA, Sastry NG, Poroikov VV. Lagunin AA, et al. PLoS One. 2018 Jan 25;13(1):e0191838. doi: 10.1371/journal.pone.0191838. eCollection 2018. PLoS One. 2018. PMID: 29370280 Free PMC article. - Connectivity mapping using a combined gene signature from multiple colorectal cancer datasets identified candidate drugs including existing chemotherapies.
Wen Q, O'Reilly P, Dunne PD, Lawler M, Van Schaeybroeck S, Salto-Tellez M, Hamilton P, Zhang SD. Wen Q, et al. BMC Syst Biol. 2015;9 Suppl 5(Suppl 5):S4. doi: 10.1186/1752-0509-9-S5-S4. Epub 2015 Sep 1. BMC Syst Biol. 2015. PMID: 26356760 Free PMC article. - A novel approach for predicting upstream regulators (PURE) that affect gene expression.
Nguyen TM, Craig DB, Tran D, Nguyen T, Draghici S. Nguyen TM, et al. Sci Rep. 2023 Oct 30;13(1):18571. doi: 10.1038/s41598-023-41374-0. Sci Rep. 2023. PMID: 37903768 Free PMC article.
References
- Smalley JL, Gant TW, Zhang SD. Application of connectivity mapping in predictive toxicology based on gene-expression similarity. Toxicology. 2010;268:143–146. - PubMed
- Lamb J. The connectivity map: a new tool for biomedical research. Nat Rev Cancer. 2007;7:54–60. - PubMed
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, et al. The connectivity map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. - PubMed
- Gullans SR. Connecting the dots using gene-expression profiles. N Engl J Med. 2006;355:2042–2044. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous