Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors - PubMed (original) (raw)
Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors
Yingpu Yu et al. Nucleic Acids Res. 2011 Jun.
Abstract
Type 2 internal ribosomal entry sites (IRESs) of encephalomyocarditis virus (EMCV), foot-and-mouth disease virus (FMDV) and other picornaviruses comprise five major domains H-L. Initiation of translation on these IRESs begins with specific binding of the central domain of initiation factor, eIF4G to the J-K domains, which is stimulated by eIF4A. eIF4G/eIF4A then restructure the region of ribosomal attachment on the IRES and promote recruitment of ribosomal 43S pre-initiation complexes. In addition to canonical translation factors, type 2 IRESs also require IRES trans-acting factors (ITAFs) that are hypothesized to stabilize the optimal IRES conformation that supports efficient ribosomal recruitment: the EMCV IRES is stimulated by pyrimidine tract binding protein (PTB), whereas the FMDV IRES requires PTB and ITAF(45). To test this hypothesis, we assessed the effect of ITAFs on the conformations of EMCV and FMDV IRESs by comparing their influence on hydroxyl radical cleavage of these IRESs from the central domain of eIF4G. The observed changes in cleavage patterns suggest that cognate ITAFs promote similar conformational changes that are consistent with adoption by the IRESs of comparable, more compact structures, in which domain J undergoes local conformational changes and is brought into closer proximity to the base of domain I.
Figures
Figure 1.
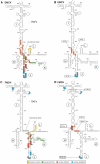
Influence of nPTB on hydroxyl radical cleavage of the EMCV IRES in IRES/eIF4Gm/eIF4A complexes from Fe(II)-tethered eIF4Gm. (A) Ribbon diagram of the HEAT-1 domain of eIF4G (PDB: 1hu3), with spheres indicating newly introduced cysteines. (B–D) Primer extension analysis of hydroxyl radical cleavage of the IRES from Fe(II) tethered eIF4Gm in IRES/eIF4Gm/eIF4A complexes in the presence/absence of nPTB. Lanes C, T, A, G depict the corresponding EMCV sequence. IRES domains and nucleotides are indicated on the left of each panel, and cleavage sites are shown on the right; boxed numbers indicate sites at which nPTB enhanced cleavage.
Figure 2.
Sites of hydroxyl radical cleavage in EMCV and FMDV IRESs from Fe(II)-tethered eIF4Gm in IRES/eIF4Gm/eIF4A complexes influenced by cognate ITAFs mapped onto the secondary structure of IRESs. Sites of hydroxyl radical cleavage from positions on eIF4Gm mapped onto the secondary structures of (A and B) the EMCV IRES and (C and D) the FMDV IRES (GenBank accession no. X00871). Models in panels B and D show only those sites at which cleavage was enhanced by nPTB or ITAF45. Nucleotide numbering of the EMCV IRES and nomenclature of IRES domains are as in (ref. 37). Cleavage sites are show in colors that match the colors of corresponding spheres in Figure 1A. Sites of strong cleavage are indicated by thick edging.
Figure 3.
Influence of nPTB and ITAF45 on hydroxyl radical cleavage of the FMDV IRES in IRES/eIF4Gm/eIF4A complexes from Fe(II)-tethered eIF4Gm. (A) Ribbon diagram of the HEAT-1 domain of eIF4Gm, with spheres indicating newly introduced cysteines. (B–E) Primer extension analysis of hydroxyl radical cleavage of the IRES from Fe(II)-tethered eIF4Gm in IRES/eIF4Gm/eIF4A complexes in the presence/absence of nPTB and ITAF45. Lanes C, T, A, G depict the corresponding FMDV sequence. IRES domains and nucleotides are indicated on the left of each panel, and cleavage sites are shown on the right; boxed numbers indicate sites at which cleavage was enhanced by nPTB and ITAF45.
Figure 4.
Comparison of changes induced by ITAFs and by 43S complexes in the pattern of hydroxyl radical cleavage of EMCV and FMDV IRESs from Fe(II) tethered eIF4Gm. Primer extension analysis of hydroxyl radical cleavage of (A–D) the EMCV IRES and (E–G) the FMDV IRES from Fe(II)-tethered eIF4Gm in IRES/eIF4Gm/eIF4A complexes in the presence of nPTB, ITAF45 or 43S complexes, as indicated. Lanes C, T, A, G depict corresponding EMCV and FMDV sequences. IRES nucleotides are indicated on the left of each panel; positions of cleaved nucleotides on the right are annotated to indicate sites of enhanced cleavage (solid boxes) and reduced cleavage (dashed box).
Figure 5.
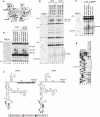
Hydroxyl radical cleavage of EMCV and FMDV IRESs from eIF4A in IRES/eIF4Gm/eIF4A ternary complexes. (A) Ribbon diagram of eIF4A in the closed ATP/RNA-bound conformation (PDB: 3EX7), with spheres indicating newly introduced cysteines. Primer extension analysis of hydroxyl radical cleavage of (B and C) the EMCV IRES and (D and E) the FMDV IRES from Fe(II)-tethered eIF4A in IRES/eIF4Gm/eIF4A complexes in the presence/absence of eIF4B, nPTB/ITAF45, ATP and adenosine 5′-[β,γ-imido] triphosphate (AMPPNP), as indicated. Lanes C, T, A, G depict corresponding EMCV and FMDV sequences. IRES nucleotides are indicated on the left of each panel, and cleavage sites are shown on the right. (F) Sites of hydroxyl radical cleavage from positions on eIF4A mapped onto the secondary structure of EMCV and FMDV IRESs. Cleavage sites are shown in colors that match the colors of corresponding spheres in (A).
Figure 6.
Hydroxyl radical cleavage of 18S rRNA from eIF4Gm in eIF4Gm-associated 43S complexes. (A and B) Primer extension analysis of hydroxyl radical cleavage of 18S rRNA from Fe(II)-tethered eIF4Gm in eIF4Gm-bound 43S complexes. Lanes C, T, A, G depict the corresponding 18S rRNA sequence. 18S rRNA nucleotides are indicated on the left of each panel, and cleavage sites are shown on the right. (C) Cleavages in 18S rRNA from eIF4Gm (cyan spheres) mapped onto the crystal structure of the yeast 80S ribosome (31). 18S rRNA and ribosomal proteins are shown as grey and blue ribbons. ES6 is colored orange and helices within it are numbered. The radius of the spheres is proportional to the efficiency of cleavage.
Similar articles
- 40S recruitment in the absence of eIF4G/4A by EMCV IRES refines the model for translation initiation on the archetype of Type II IRESs.
Chamond N, Deforges J, Ulryck N, Sargueil B. Chamond N, et al. Nucleic Acids Res. 2014;42(16):10373-84. doi: 10.1093/nar/gku720. Epub 2014 Aug 26. Nucleic Acids Res. 2014. PMID: 25159618 Free PMC article. - Initiation on the divergent Type I cadicivirus IRES: factor requirements and interactions with the translation apparatus.
Asnani M, Pestova TV, Hellen CU. Asnani M, et al. Nucleic Acids Res. 2016 Apr 20;44(7):3390-407. doi: 10.1093/nar/gkw074. Epub 2016 Feb 11. Nucleic Acids Res. 2016. PMID: 26873921 Free PMC article. - In vitro reconstitution and biochemical characterization of translation initiation by internal ribosomal entry.
Kolupaeva VG, de Breyne S, Pestova TV, Hellen CU. Kolupaeva VG, et al. Methods Enzymol. 2007;430:409-39. doi: 10.1016/S0076-6879(07)30016-5. Methods Enzymol. 2007. PMID: 17913647 - Advances and Breakthroughs in IRES-Directed Translation and Replication of Picornaviruses.
Abdullah SW, Wu J, Wang X, Guo H, Sun S. Abdullah SW, et al. mBio. 2023 Apr 25;14(2):e0035823. doi: 10.1128/mbio.00358-23. Epub 2023 Mar 20. mBio. 2023. PMID: 36939331 Free PMC article. Review. - Translation of encephalomyocarditis virus RNA by internal ribosomal entry.
Hellen CU, Wimmer E. Hellen CU, et al. Curr Top Microbiol Immunol. 1995;203:31-63. doi: 10.1007/978-3-642-79663-0_2. Curr Top Microbiol Immunol. 1995. PMID: 7555090 Review.
Cited by
- N-terminal domain of polypyrimidine-tract binding protein is a dynamic folding platform for adaptive RNA recognition.
Damberger FF, Krepl M, Arora R, Beusch I, Maris C, Dorn G, Šponer J, Ravindranathan S, Allain FH. Damberger FF, et al. Nucleic Acids Res. 2024 Sep 23;52(17):10683-10704. doi: 10.1093/nar/gkae713. Nucleic Acids Res. 2024. PMID: 39180402 Free PMC article. - Ribosomal Protein L13 Promotes IRES-Driven Translation of Foot-and-Mouth Disease Virus in a Helicase DDX3-Dependent Manner.
Han S, Sun S, Li P, Liu Q, Zhang Z, Dong H, Sun M, Wu W, Wang X, Guo H. Han S, et al. J Virol. 2020 Jan 6;94(2):e01679-19. doi: 10.1128/JVI.01679-19. Print 2020 Jan 6. J Virol. 2020. PMID: 31619563 Free PMC article. - The mechanism of translation initiation on Aichivirus RNA mediated by a novel type of picornavirus IRES.
Yu Y, Sweeney TR, Kafasla P, Jackson RJ, Pestova TV, Hellen CU. Yu Y, et al. EMBO J. 2011 Aug 26;30(21):4423-36. doi: 10.1038/emboj.2011.306. EMBO J. 2011. PMID: 21873976 Free PMC article. - Host factors in enterovirus 71 replication.
Shih SR, Stollar V, Li ML. Shih SR, et al. J Virol. 2011 Oct;85(19):9658-66. doi: 10.1128/JVI.05063-11. Epub 2011 Jun 29. J Virol. 2011. PMID: 21715481 Free PMC article. Review. - Biological function of Foot-and-mouth disease virus non-structural proteins and non-coding elements.
Gao Y, Sun SQ, Guo HC. Gao Y, et al. Virol J. 2016 Jun 22;13:107. doi: 10.1186/s12985-016-0561-z. Virol J. 2016. PMID: 27334704 Free PMC article. Review.
References
- Pestova TV, Borukhov SI, Hellen CUT. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K01 CA119107/CA/NCI NIH HHS/United States
- R01 AI051340/AI/NIAID NIH HHS/United States
- AI51340/AI/NIAID NIH HHS/United States
- R01 GM095720/GM/NIGMS NIH HHS/United States
- GM59660/GM/NIGMS NIH HHS/United States
- R56 AI051340/AI/NIAID NIH HHS/United States
- R01 AI051340-09/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous