Discovery of a small arterivirus gene that overlaps the GP5 coding sequence and is important for virus production - PubMed (original) (raw)
Discovery of a small arterivirus gene that overlaps the GP5 coding sequence and is important for virus production
Andrew E Firth et al. J Gen Virol. 2011 May.
Abstract
The arterivirus family (order Nidovirales) of single-stranded, positive-sense RNA viruses includes porcine respiratory and reproductive syndrome virus and equine arteritis virus (EAV). Their replicative enzymes are translated from their genomic RNA, while their seven structural proteins are encoded by a set of small, partially overlapping genes in the genomic 3'-proximal region. The latter are expressed via synthesis of a set of subgenomic mRNAs that, in general, are functionally monocistronic (except for a bicistronic mRNA encoding the E and GP2 proteins). ORF5, which encodes the major glycoprotein GP5, has been used extensively for phylogenetic analyses. However, an in-depth computational analysis now reveals the arterivirus-wide conservation of an additional AUG-initiated ORF, here termed ORF5a, that overlaps the 5' end of ORF5. The pattern of substitutions across sequence alignments indicated that ORF5a is subject to functional constraints at the amino acid level, while an analysis of substitutions at synonymous sites in ORF5 revealed a greatly reduced frequency of substitution in the portion of ORF5 that is overlapped by ORF5a. The 43-64 aa ORF5a protein and GP5 are probably expressed from the same subgenomic mRNA, via a translation initiation mechanism involving leaky ribosomal scanning. Inactivation of ORF5a expression by reverse genetics yielded a severely crippled EAV mutant, which displayed lower titres and a tiny plaque phenotype. These defects, which could be partially complemented in ORF5a-expressing cells, indicate that the novel protein, which may be the eighth structural protein of arteriviruses, is expressed and important for arterivirus infection.
Figures
Fig. 1.
(a) EAV genome organization and expression. The positions of the ORFs encoding the seven known structural proteins and the newly discovered ORF5a are indicated. ORF5a slightly overlaps with the GP4 gene and largely overlaps with the GP5 gene. Small grey boxes indicate the positions of the TRS that direct sg RNA synthesis and serve as junction sequences for the 5′ common leader sequence of arterivirus mRNAs. (b) Maps of the ORF5a/ORF5 region of the genomes of representative arterivirus sequences (see Fig. 2b for GenBank accession numbers). For EAV and the two PRRSV genotypes, plots are shown for several coding-potential statistics in a 15-codon sliding window. The top and middle panels illustrate the degree of conservation at synonymous sites within the GP5 CDS: the top panel depicts the probability that the degree of conservation within a given window could be obtained under a null model of neutral evolution at synonymous sites, while the middle panel depicts the absolute amount of conservation as represented by the ratio of the observed number of substitutions within a given window to the number expected under the null model. Scores below the dashed line correspond to more conserved regions and are indicative of overlapping functional elements, either coding or non-coding. Owing to the huge quantity of GP5 sequence data (8344 sequences) available for the PRRSV-NA plot, the formal P values for this analysis are extremely small for the conserved 5′ region. The bottom panel depicts the
mlogd
coding potential score in the +2 frame (relative to the GP5 CDS). For illustrative purposes the M CDS, and the GP4 CDS in PRRSV-NA, were not included in the null-model CDS annotations so that these CDSs, like ORF5a, register positive coding potential (i.e. scores above the dashed line) in the
mlogd
analysis. Note that, regardless of sign (positive or negative), the
mlogd
signal tends to be weaker within regions of high conservation (e.g. within ORF5a) due to there being fewer substitutions with which to discriminate between the null model and the alternative model. A dashed vertical line indicates the 3′ end of the common ORF5a variant (46 codons instead of 51 codons) in PRRSV-NA.
Fig. 2.
Expression of arterivirus ORF5a. (a) RNA sequence and translation of the EAV ORF5a region, showing the slightly overlapping GP4 and GP5 genes, and the newly discovered ORF5a that overlaps with both. The TRS for sg RNA5 synthesis and relevant initiation and termination codons are indicated. Red underscores indicate residues mutated to knock out ORF5a in mutant Δ5a-2. (b) Context of the ORF5a (green) and ORF5 (blue) translation initiation codons in five representative arterivirus sequences, illustrating the potential for leaky scanning being the translation initiation mechanism for the second reading frame (see text for details). The figure shows that the ORF5a initiation codon is the more upstream in EAV and PRRSV-NA and the more downstream in the other arteriviruses. The ORF4 termination codon is boxed and the two residues mutated to knock out EAV ORF5a in mutant Δ5a-2 are indicated by arrows. (c) Amino acid sequences of the ORF5a proteins of five representative arterivirus sequences. The position of the putative transmembrane domain, as predicted by
phobius
(
; Käll et al., 2004), is indicated in yellow. The ORF5a protein is predicted to be a type III membrane protein (lumenal amino-terminal domain, central signal-anchor/transmembrane domain and cytoplasmic carboxy-terminal domain).
Fig. 3.
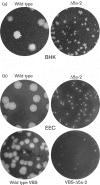
(a) Comparison of plaque morphology on BHK-21 cells of wild-type EAV and ORF5a knockout mutant Δ5a-2, illustrating the severely crippled phenotype of the latter. In addition to the dramatically reduced plaque size, Δ5a-2 infectivity titres were reduced by approximately two logs. (b) Comparison of wild-type and Δ5a-2 plaque morphology on equine endothelial (EEC) cells using both a cell-culture-adapted EAV isolate (EAV515) and an isolate that is virulent in horses (VBS). In both virus backbones, Δ5a-2 plaque sizes and titres are severely reduced compared with those of the corresponding parental virus.
Fig. 4.
Complementation of the EAV ORF5a knockout mutant Δ5a-2 in ORF5a-expressing cells. The Δ5a-2 small-plaque phenotype (left) was partially complemented when the plaque assay was performed using BHK-21 cells carrying an ORF5a-expressing SINV RNA replicon (middle; Agapov et al., 1998). As a negative control, cells carrying a GFP-expressing replicon were used. The plaque morphology of wild-type virus on ORF5a-expressing cells is shown on the right.
Similar articles
- Arterivirus minor envelope proteins are a major determinant of viral tropism in cell culture.
Tian D, Wei Z, Zevenhoven-Dobbe JC, Liu R, Tong G, Snijder EJ, Yuan S. Tian D, et al. J Virol. 2012 Apr;86(7):3701-12. doi: 10.1128/JVI.06836-11. Epub 2012 Jan 18. J Virol. 2012. PMID: 22258262 Free PMC article. - Identification of a novel structural protein of arteriviruses.
Snijder EJ, van Tol H, Pedersen KW, Raamsman MJ, de Vries AA. Snijder EJ, et al. J Virol. 1999 Aug;73(8):6335-45. doi: 10.1128/JVI.73.8.6335-6345.1999. J Virol. 1999. PMID: 10400725 Free PMC article. - Novel structural protein in porcine reproductive and respiratory syndrome virus encoded by an alternative ORF5 present in all arteriviruses.
Johnson CR, Griggs TF, Gnanandarajah J, Murtaugh MP. Johnson CR, et al. J Gen Virol. 2011 May;92(Pt 5):1107-1116. doi: 10.1099/vir.0.030213-0. Epub 2011 Feb 9. J Gen Virol. 2011. PMID: 21307222 Free PMC article. - Membrane proteins of arterivirus particles: structure, topology, processing and function.
Veit M, Matczuk AK, Sinhadri BC, Krause E, Thaa B. Veit M, et al. Virus Res. 2014 Dec 19;194:16-36. doi: 10.1016/j.virusres.2014.09.010. Epub 2014 Sep 30. Virus Res. 2014. PMID: 25278143 Free PMC article. Review. - Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: comparison of the North American and European isolates.
Dea S, Gagnon CA, Mardassi H, Pirzadeh B, Rogan D. Dea S, et al. Arch Virol. 2000;145(4):659-88. doi: 10.1007/s007050050662. Arch Virol. 2000. PMID: 10893147 Free PMC article. Review.
Cited by
- Expression and Antibody Preparation of GP5a Gene of Porcine Reproductive and Respiratory Syndrome Virus.
Wei C, Huang Z, Sun L, Xie J, Chen Y, Zhang M, Zhang C, Qi H, Qi W, Ning Z, Yuan L, Wang H, Zhang L, Zhang G. Wei C, et al. Indian J Microbiol. 2013 Sep;53(3):370-5. doi: 10.1007/s12088-013-0368-1. Epub 2013 Mar 2. Indian J Microbiol. 2013. PMID: 24426138 Free PMC article. - Identification of the strain-specifically truncated nonstructural protein 10 of porcine reproductive and respiratory syndrome virus in infected cells.
Zhang ZB, Xu L, Wen XX, Dong JG, Zhou L, Ge XN, Yang HC, Guo X. Zhang ZB, et al. J Integr Agric. 2018 May;17(5):1171-1180. doi: 10.1016/S2095-3119(17)61896-3. Epub 2018 May 11. J Integr Agric. 2018. PMID: 32288956 Free PMC article. - Virus replicon particles expressing porcine reproductive and respiratory syndrome virus proteins elicit immune priming but do not confer protection from viremia in pigs.
Eck M, Durán MG, Ricklin ME, Locher S, Sarraseca J, Rodríguez MJ, McCullough KC, Summerfield A, Zimmer G, Ruggli N. Eck M, et al. Vet Res. 2016 Feb 19;47:33. doi: 10.1186/s13567-016-0318-0. Vet Res. 2016. PMID: 26895704 Free PMC article. - Interplay between interferon-mediated innate immunity and porcine reproductive and respiratory syndrome virus.
Sun Y, Han M, Kim C, Calvert JG, Yoo D. Sun Y, et al. Viruses. 2012 Apr;4(4):424-46. doi: 10.3390/v4040424. Epub 2012 Apr 2. Viruses. 2012. PMID: 22590680 Free PMC article. Review. - Characterization and Pathogenicity of Two Novel PRRSVs Recombined by NADC30-like and NADC34-like Strains in China.
Wu Y, Peng O, Xu Q, Li Q, Li W, Lin L, Zhou Q, Cai X, Hu G, He Z, Chen Y, Zhang H. Wu Y, et al. Viruses. 2022 Sep 30;14(10):2174. doi: 10.3390/v14102174. Viruses. 2022. PMID: 36298730 Free PMC article.
References
- Allende R., Lewis T. L., Lu Z., Rock D. L., Kutish G. F., Ali A., Doster A. R., Osorio F. A. (1999). North American and European porcine reproductive and respiratory syndrome viruses differ in non-structural protein coding regions. J Gen Virol 80, 307–315 - PubMed
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J Mol Biol 215, 403–410 - PubMed
- Balasuriya U. B., Snijder E. J., van Dinten L. C., Heidner H. W., Wilson W. D., Hedges J. F., Hullinger P. J., MacLachlan N. J. (1999). Equine arteritis virus derived from an infectious cDNA clone is attenuated and genetically stable in infected stallions. Virology 260, 201–208 10.1006/viro.1999.9817 - DOI - PubMed
- Balasuriya U. B., Snijder E. J., Heidner H. W., Zhang J., Zevenhoven-Dobbe J. C., Boone J. D., McCollum W. H., Timoney P. J., MacLachlan N. J. (2007). Development and characterization of an infectious cDNA clone of the virulent Bucyrus strain of Equine arteritis virus. J Gen Virol 88, 918–924 10.1099/vir.0.82415-0 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous