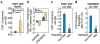9p21 DNA variants associated with coronary artery disease impair interferon-γ signalling response - PubMed (original) (raw)
9p21 DNA variants associated with coronary artery disease impair interferon-γ signalling response
Olivier Harismendy et al. Nature. 2011.
Abstract
Genome-wide association studies have identified single nucleotide polymorphisms (SNPs) in the 9p21 gene desert associated with coronary artery disease (CAD) and type 2 diabetes. Despite evidence for a role of the associated interval in neighbouring gene regulation, the biological underpinnings of these genetic associations with CAD or type 2 diabetes have not yet been explained. Here we identify 33 enhancers in 9p21; the interval is the second densest gene desert for predicted enhancers and six times denser than the whole genome (P < 6.55 × 10(-33)). The CAD risk alleles of SNPs rs10811656 and rs10757278 are located in one of these enhancers and disrupt a binding site for STAT1. Lymphoblastoid cell lines homozygous for the CAD risk haplotype show no binding of STAT1, and in lymphoblastoid cell lines homozygous for the CAD non-risk haplotype, binding of STAT1 inhibits CDKN2BAS (also known as CDKN2B-AS1) expression, which is reversed by short interfering RNA knockdown of STAT1. Using a new, open-ended approach to detect long-distance interactions, we find that in human vascular endothelial cells the enhancer interval containing the CAD locus physically interacts with the CDKN2A/B locus, the MTAP gene and an interval downstream of IFNA21. In human vascular endothelial cells, interferon-γ activation strongly affects the structure of the chromatin and the transcriptional regulation in the 9p21 locus, including STAT1-binding, long-range enhancer interactions and altered expression of neighbouring genes. Our findings establish a link between CAD genetic susceptibility and the response to inflammatory signalling in a vascular cell type and thus demonstrate the utility of genome-wide association study findings in directing studies to novel genomic loci and biological processes important for disease aetiology.
Conflict of interest statement
Author Information The authors declare no competing interests
Figures
Figure 1. Functional annotation of the 9p21 interval
The locations of the core CAD and T2D associated intervals (track A) and the predicted insulators (brown), enhancers (orange) and promoters (green) in HeLa cells are indicated (track B). The enhancers are distributed (track C) between the CAD interval (red), the T2D interval (blue) or located outside (orange). The location of the binding sites for FoxA1 in MCF7 cells (track D2) and STAT1 in IFNγ treated and non-treated HeLa (track D3) as well as the distribution of 9p21 chromatin marks in the ENCODE data (tracks E1 and E2 – Supplemental Methods) are indicated.
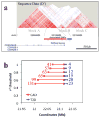
Figure 2. LD analysis of the 9p21 interval
(a) The sequenced interval shows three LD blocks. The 53-kb CAD interval is located at the 3′ end of the first block (A), the second block (B) spans 11-kb corresponding to the T2D interval and the third block (C) spans 63-kb. LD map (D′) based on variants identified in the 50 sequenced samples. (b) Number of variants in LD at various _r_2 thresholds (y axis) with any CAD (red) or T2D associated variants (blue). The distance spanned by the SNPs in LD is indicated (x axis).
Figure 3. In vivo effects of the ECAD9 variants
(a) Enrichment of the ECAD9 STAT1 binding site by anti-STAT1 ChIP in HUVEC cells untreated or treated with IFNγ. (b) Changes in level of expression of CDKN2B and CDKN2BAS genes upon treatment with IFNγ in HeLa and HUVEC. (c) Enrichment of the ECAD9 STAT1 binding site by anti-STAT1 ChIP in LCL homozygous for the CAD non-risk or CAD risk haplotypes. (d) Expression level changes of CDKN2BAS in LCL homozygous for non-risk or risk CAD haplotype after STAT1 knock-down by siRNA. (*) and (**) indicate two-tailed Student T-test p <0.05 and p<0.01, respectively.
Figure 4. Long-Range interaction with the enhancer locus
(a) Circular representation of the 3D-DSL results. The scale shows chromosome 9 position (hg18) in 100kb increments. RefSeq genes (dark blue) and HeLa predicted enhancers (orange) are displayed with the CAD (red) and T2D (blue) associated enhancers. The histogram represents the normalized average number of reads mapping to each BamH1 donor site. The inner circle links connect BamH1 acceptor sites for the 9 most strongly interacting donor sites. (b) PCR validation of the long-range interaction between ECAD9 and CDKN2A/B (upper panel) or MTAP (lower panel). Arrow indicates the specific product.
Comment in
- Complex disease: Finding functions in the wilderness.
Skipper M. Skipper M. Nat Rev Genet. 2011 Mar;12(3):153. doi: 10.1038/nrg2962. Nat Rev Genet. 2011. PMID: 21331085 No abstract available. - Regulatory elements in noncoding DNA in the chromosome 9p21 locus.
Musunuru K. Musunuru K. Circ Cardiovasc Genet. 2011 Jun;4(3):330-1. doi: 10.1161/CIRCGENETICS.111.960500. Circ Cardiovasc Genet. 2011. PMID: 21673313 No abstract available. - Enduring mystery of the chromosome 9p21.3 locus.
Musunuru K. Musunuru K. Circ Cardiovasc Genet. 2013 Apr;6(2):224-5. doi: 10.1161/CIRCGENETICS.113.000132. Circ Cardiovasc Genet. 2013. PMID: 23591041 No abstract available.
Similar articles
- Interferon-γ activates expression of p15 and p16 regardless of 9p21.3 coronary artery disease risk genotype.
Almontashiri NA, Fan M, Cheng BL, Chen HH, Roberts R, Stewart AF. Almontashiri NA, et al. J Am Coll Cardiol. 2013 Jan 15;61(2):143-7. doi: 10.1016/j.jacc.2012.08.1020. Epub 2012 Nov 28. J Am Coll Cardiol. 2013. PMID: 23199516 - Common SNP-based haplotype analysis of the 9p21.3 gene locus as predictor coronary artery disease in Tanzanian population.
Akan G, Kisenge P, Sanga TS, Mbugi E, Adolf I, Turkcan MK, Janabi M, Atalar F. Akan G, et al. Cell Mol Biol (Noisy-le-grand). 2019 Jul 31;65(6):33-43. Cell Mol Biol (Noisy-le-grand). 2019. PMID: 31472045 - Haplotypes on 9p21 modify the risk for coronary artery disease among Indians.
AshokKumar M, Emmanuel C, Dhandapany PS, Rani DS, SaiBabu R, Cherian KM, Thangaraj K. AshokKumar M, et al. DNA Cell Biol. 2011 Feb;30(2):105-10. doi: 10.1089/dna.2010.1046. Epub 2010 Sep 21. DNA Cell Biol. 2011. PMID: 20858033 - Functional genomics of the CDKN2A/B locus in cardiovascular and metabolic disease: what have we learned from GWASs?
Hannou SA, Wouters K, Paumelle R, Staels B. Hannou SA, et al. Trends Endocrinol Metab. 2015 Apr;26(4):176-84. doi: 10.1016/j.tem.2015.01.008. Epub 2015 Mar 3. Trends Endocrinol Metab. 2015. PMID: 25744911 Review. - Functional genomics of the 9p21.3 locus for atherosclerosis: clarity or confusion?
Chen HH, Almontashiri NA, Antoine D, Stewart AF. Chen HH, et al. Curr Cardiol Rep. 2014 Jul;16(7):502. doi: 10.1007/s11886-014-0502-7. Curr Cardiol Rep. 2014. PMID: 24893939 Review.
Cited by
- An integrated encyclopedia of DNA elements in the human genome.
ENCODE Project Consortium. ENCODE Project Consortium. Nature. 2012 Sep 6;489(7414):57-74. doi: 10.1038/nature11247. Nature. 2012. PMID: 22955616 Free PMC article. - Final "perspectives on the news": American Association of Clinical Endocrinology and American Diabetes Association 2011.
Bloomgarden ZT. Bloomgarden ZT. Diabetes Care. 2011 Dec;34(12):e176-81. doi: 10.2337/dc11-1800. Diabetes Care. 2011. PMID: 22110173 Free PMC article. No abstract available. - Functional anatomy of distant-acting mammalian enhancers.
Dickel DE, Visel A, Pennacchio LA. Dickel DE, et al. Philos Trans R Soc Lond B Biol Sci. 2013 May 6;368(1620):20120359. doi: 10.1098/rstb.2012.0359. Print 2013. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23650633 Free PMC article. Review. - Epigenetic regulation in human melanoma: past and future.
Sarkar D, Leung EY, Baguley BC, Finlay GJ, Askarian-Amiri ME. Sarkar D, et al. Epigenetics. 2015;10(2):103-21. doi: 10.1080/15592294.2014.1003746. Epigenetics. 2015. PMID: 25587943 Free PMC article. Review. - A Functional Indel Polymorphism Within MIR155HG Is Associated With Sudden Cardiac Death Risk in a Chinese Population.
Zhang Q, Yu H, Yang Z, Li L, He Y, Zhu S, Li C, Zhang S, Luo B, Gao Y. Zhang Q, et al. Front Cardiovasc Med. 2021 May 31;8:671168. doi: 10.3389/fcvm.2021.671168. eCollection 2021. Front Cardiovasc Med. 2021. PMID: 34136547 Free PMC article.
References
- Helgadottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–3. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK039949-29/DK/NIDDK NIH HHS/United States
- R01 NS034934/NS/NINDS NIH HHS/United States
- DK018477/DK/NIDDK NIH HHS/United States
- R37 DK039949/DK/NIDDK NIH HHS/United States
- UL1 RR025774/RR/NCRR NIH HHS/United States
- U01 HL107442/HL/NHLBI NIH HHS/United States
- R01 DK018477-35/DK/NIDDK NIH HHS/United States
- DK74686/DK/NIDDK NIH HHS/United States
- R01 CA097134/CA/NCI NIH HHS/United States
- R01 HL065445-12/HL/NHLBI NIH HHS/United States
- R01 DK018477/DK/NIDDK NIH HHS/United States
- R21 CA152613-01/CA/NCI NIH HHS/United States
- 1U54RR025204/RR/NCRR NIH HHS/United States
- 1UL1RR031980-01/RR/NCRR NIH HHS/United States
- NS34934/NS/NINDS NIH HHS/United States
- 1UL1RR025774/RR/NCRR NIH HHS/United States
- P01 AG025204-01/AG/NIA NIH HHS/United States
- L65445/PHS HHS/United States
- P01 DK074868/DK/NIDDK NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- 1R21CA152613-01/CA/NCI NIH HHS/United States
- HL065445/HL/NHLBI NIH HHS/United States
- R01 HL065445/HL/NHLBI NIH HHS/United States
- UL1 RR031980-01/RR/NCRR NIH HHS/United States
- DK39949/DK/NIDDK NIH HHS/United States
- R21 CA152613/CA/NCI NIH HHS/United States
- R01 DK039949/DK/NIDDK NIH HHS/United States
- UL1 RR025774-01/RR/NCRR NIH HHS/United States
- UL1 RR031980/RR/NCRR NIH HHS/United States
- DK074868/DK/NIDDK NIH HHS/United States
- CA97134/CA/NCI NIH HHS/United States
- R21 CA152613-02/CA/NCI NIH HHS/United States
- P01 AG025204/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous