Activity-induced Notch signaling in neurons requires Arc/Arg3.1 and is essential for synaptic plasticity in hippocampal networks - PubMed (original) (raw)
Comparative Study
. 2011 Feb 10;69(3):437-44.
doi: 10.1016/j.neuron.2011.01.004.
Shuxi Liu, Yue Wang, Ramy Badie, Constance Smith-Hicks, Jing Wu, Tarran J Pierfelice, Bagrat Abazyan, Mark P Mattson, Dietmar Kuhl, Mikhail Pletnikov, Paul F Worley, Nicholas Gaiano
Affiliations
- PMID: 21315255
- PMCID: PMC3056341
- DOI: 10.1016/j.neuron.2011.01.004
Comparative Study
Activity-induced Notch signaling in neurons requires Arc/Arg3.1 and is essential for synaptic plasticity in hippocampal networks
Lavinia Alberi et al. Neuron. 2011.
Abstract
Notch signaling in the nervous system has been most studied in the context of cell fate specification. However, numerous studies have suggested that Notch also regulates neuronal morphology, synaptic plasticity, learning, and memory. Here we show that Notch1 and its ligand Jagged1 are present at the synapse, and that Notch signaling in neurons occurs in response to synaptic activity. In addition, neuronal Notch signaling is positively regulated by Arc/Arg3.1, an activity-induced gene required for synaptic plasticity. In Arc/Arg3.1 mutant neurons, the proteolytic activation of Notch1 is disrupted both in vivo and in vitro. Conditional deletion of Notch1 in the postnatal hippocampus disrupted both long-term potentiation (LTP) and long-term depression (LTD), and led to deficits in learning and short-term memory. Thus, Notch signaling is dynamically regulated in response to neuronal activity, Arc/Arg3.1 is a context-dependent Notch regulator, and Notch1 is required for the synaptic plasticity that contributes to memory formation.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
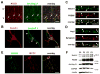
Figure 1. Notch1 is present at the synapse in mature neurons
(A) The somatosensory cortex is shown. Arc and NICD1 are both present in the soma (arrows) and apical dendrites of layer V neurons. (B) DIV21 (21 days in vitro) hippocampal neuronal cultures immunostained to detect Notch1 and Jag1. While Notch1 is localized to the cell soma and dendrites, the ligand Jag1 is enriched in axonal processes (arrowheads). (C) Notch1 co-localizes in dendritic spines with the synaptic protein PSD95 (see also Figure S1). (D) Jag1 co-localizes with the synaptic protein Synapsin I. (E) The activated form of Notch-1 (NICD1) co-localizes with PSD95 in DIV21 neurons. (F) Subcellular fractionation of adult mouse cerebral cortical tissue reveals that S3 fragment of Notch1 (asterisk) is enriched in synaptosomal fractions (P2, washed P2′, and membranes P3), as compared to the cytoplasmic fraction (S2). Scale bars=75 μm (A), 25 μm (B,E), 5 μm (C,D).
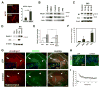
Figure 2. Notch signaling occurs in neurons in response to activity
(A) NICD1 is increased in cultured hippocampal neurons after NMDA treatment. Relative NICD1 signal levels with (n=4) or without (n=3) treatment (right). Scale bar=40 μm. (B) Western blot analysis showing that NMDA increased NICD1 (2.8-fold, n=3, p<0.05) and Arc protein levels (6.6-fold, n=3, p<0.01), while NMDA receptor blockade (AP5) decreased NICD1 (4.3-fold, n=3, p<0.05), and Arc (10.0-fold, n=3, p<0.01). EDTA treatment was used a positive control to activate Notch1 (Rand et al., 2000). (C) Treatment of hippocampal neurons with bicuculline increased Arc and Notch1 S3 fragment (2.9-fold at 4 hours, n=5, p<0.001) levels (asterisk). (D) Western blot (WB) showing that full-length (pre-S1 cleavage) Notch1 protein levels increase in response to bicuculline, even with the transcriptional inhibitor actinomycin-D (Act-D). (E) Quantification of four experiments shows that the expression of full length Notch1 (normalized to β-actin) is substantially increased after bicuculline treatment, with or without Act-D. ns, not significant. S.D. is shown. β-tub, β-tubulin, βact, β-actin. (F) Quantitative RT-PCR of hippocampal cultures treated with bicuculline for 4 hours shows that Jag1 expression was increased in response to increased neuronal activity, *p<0.04. n=4. (G) 3.5 hours after LTP induction in the CA1 region of acute hippocampal slices from adult mice, both Arc and Notch1 protein levels were elevated in the soma of CA3 (arrow) and CA1 (arrowhead) neurons. (H) IHC revealed increased NICD1 in CA1 in response to LTP. (I) Plot of field excitatory post-synaptic potential (fEPSP) in hippocampal slices. Mean values from four animal are shown. Scale bars=50μm.
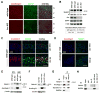
Figure 3. Neuronal Notch signaling is disrupted in Arc mutants in vivo
(A) Five week old Arc mutant and wild type animals containing the TNR transgene were examined to determine the impact of Arc disruption on the Notch pathway in vivo. Arc mutants had reduced EGFP expression, indicating reduced Notch activation (somatosensory cortex is shown). (B) WB of cell lysates derived from the cerebral cortex of Arc knockout (KO) animals revealed reduced NICD1 generation (asterisk) and EGFP expression. Two different exposures of the S3 band are shown. NICD1 band intensity (normalized to β-tubulin) was compared between 6 wild type and 6 Arc KO animals. **p<0.01. S.D. is shown. Scale bar=70 μm. (C) In five week old wild type animals, exploration of a novel environment resulted in a rapid increase in Arc and Notch1 expression in CA1 (not shown) and CA3 (Notch1 signal intensity for cage control and 45 minutes after exploration was 6.8 ± 2.6 arbitrary units (a.u.), and 21.5 ± 5.2 a.u., respectively. n=3 each, p<0.01). Much of the Notch1 protein was in the cell soma and nucleus, consistent with active Notch1 signaling. (D) No increase in Notch1 expression was observed in Arc/Arc3.1 knockout (KO) animals after exploration (cage control and 45 minutes after exploration was 11.2 ± 3.3 arbitrary units (a.u.), and 13.4 ± 4.8 a.u., respectively, n=3 each). (E) Western blot analysis from Arc KO and wild-type hippocampal neuronal cultures revealed that, in absence of Arc, Notch processing is reduced; the S3 band (asterisk) is nearly absent, unlike the S1 band (upper). (F) WB of Arc mutant hippocampal cultures infected with Sindbis virus expressing either full-length Arc, or a non-functional form lacking residues 91–100 (Δ)(Chowdhury et al., 2006). (G) Arc and Dynamin co-IP with Notch-1 from cortical protein preparations. (H) Notch1 co-IPs with Arc from protein lysates generated from neuronal cultures. This interaction was not detected in Arc KO cultures. Scale bars=50 μm.
Figure 4. Loss of Notch function in CA1 affects neuronal morphology and plasticity
(A) IHC shows that in Notch1 cKO mice Notch1 expression is reduced in the CA1 region of the hippocampus (arrowheads). cKO animals had increased Notch1 expression in astrocytes (arrow). Inset scale bar=25 μm. Note that these mice were exposed to a novel environment to increase Notch1 expression. (B) Golgi impregnated CA1 pyramidal neurons reveal no difference in gross dendritic morphology between Notch1 cKO and controls. (C) Notch1 cKO CA1 neurons have comparable lengths of apical and basal dendrites. (D) In Notch1 cKO spine density of CA1 dendrites is reduced 16% (p<0.001). (E) In Notch1 cKO the number of mushroom spines is 25% reduced on CA1 pyramidal dendrites, and the number of thin spines is 40% increased (p<0.001). (F) Images of Golgi stained control and Notch1 cKO dendritic spines. Scale bar in F=10 μm. (G) Notch1 cKO (closed circles) has normal basal transmission as compared to controls (open circles). (H) Paired pulse facilitation (PPF) occurs in the Notch1 cKO mice. (I,J) LTP and LTD were reduced in the Notch1 cKO slices (p<0.01 each).
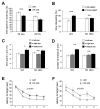
Figure 5. Notch1 conditional ablation causes deficits in memory acquisition
(A) While both control and Notch1 cKO animals spent more time exploring a novel object 25 min after exposure to two identical objects (83.2 ± 3.5% preference versus 68.8 ± 3.9%, respectively, p<0.001), 24 hours later, Notch1 cKO do not display any preference for the novel object (52.8 ± 4.5%, p = 0.4) in contrast to controls (65.4 ± 3.6%, p<0.01)(n=12 for both Notch1 cKO and wt). (B) Notch1 cKO showed no preference for a novel subject (48.3 ± 4.1%, p=0.6) in contrast to controls (60.2 ± 3.8%, p<0.01)(n=13 Notch1 cKO and n=16 control). (C,D) Despite normal alternation in a Y-maze, Notch1 cKO mice fail to show a robust preference for a previously hidden arm, while controls do (n=12 Notch1 cKO, p<0.07; n=15 control; p<0.022). (E,F) The average time to find the platform in a Morris water maze is higher on day two, three, and four for Notch1 cKO animals (p<0.05 for each time point). Similar results were obtained with reversal learning, where the platform was placed in the opposite quadrant (F). A repeated measure ANOVA was used to assess statistical significance in E and F. Twenty-four hours after the last of five training sessions for both initial and reversal learning, both Notch1 cKO and controls spend more time in the target quadrant, indicating comparable memory retrieval after repetitive learning (n=14 Notch1 cKO and n=18 wt)(Figure S6A,B).
Similar articles
- Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories.
Plath N, Ohana O, Dammermann B, Errington ML, Schmitz D, Gross C, Mao X, Engelsberg A, Mahlke C, Welzl H, Kobalz U, Stawrakakis A, Fernandez E, Waltereit R, Bick-Sander A, Therstappen E, Cooke SF, Blanquet V, Wurst W, Salmen B, Bösl MR, Lipp HP, Grant SG, Bliss TV, Wolfer DP, Kuhl D. Plath N, et al. Neuron. 2006 Nov 9;52(3):437-44. doi: 10.1016/j.neuron.2006.08.024. Neuron. 2006. PMID: 17088210 - Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors.
Shepherd JD, Rumbaugh G, Wu J, Chowdhury S, Plath N, Kuhl D, Huganir RL, Worley PF. Shepherd JD, et al. Neuron. 2006 Nov 9;52(3):475-84. doi: 10.1016/j.neuron.2006.08.034. Neuron. 2006. PMID: 17088213 Free PMC article. - The Immediate Early Gene Arc Is Not Required for Hippocampal Long-Term Potentiation.
Kyrke-Smith M, Volk LJ, Cooke SF, Bear MF, Huganir RL, Shepherd JD. Kyrke-Smith M, et al. J Neurosci. 2021 May 12;41(19):4202-4211. doi: 10.1523/JNEUROSCI.0008-20.2021. Epub 2021 Apr 8. J Neurosci. 2021. PMID: 33833081 Free PMC article. - Arc/Arg3.1: linking gene expression to synaptic plasticity and memory.
Tzingounis AV, Nicoll RA. Tzingounis AV, et al. Neuron. 2006 Nov 9;52(3):403-7. doi: 10.1016/j.neuron.2006.10.016. Neuron. 2006. PMID: 17088207 Review. - Inverse synaptic tagging: An inactive synapse-specific mechanism to capture activity-induced Arc/arg3.1 and to locally regulate spatial distribution of synaptic weights.
Okuno H, Minatohara K, Bito H. Okuno H, et al. Semin Cell Dev Biol. 2018 May;77:43-50. doi: 10.1016/j.semcdb.2017.09.025. Epub 2017 Sep 23. Semin Cell Dev Biol. 2018. PMID: 28939038 Review.
Cited by
- Group 1 metabotropic glutamate receptor function and its regulation of learning and memory in the aging brain.
Ménard C, Quirion R. Ménard C, et al. Front Pharmacol. 2012 Oct 12;3:182. doi: 10.3389/fphar.2012.00182. eCollection 2012. Front Pharmacol. 2012. PMID: 23091460 Free PMC article. - LIN-12/Notch signaling instructs postsynaptic muscle arm development by regulating UNC-40/DCC and MADD-2 in Caenorhabditis elegans.
Li P, Collins KM, Koelle MR, Shen K. Li P, et al. Elife. 2013 Mar 19;2:e00378. doi: 10.7554/eLife.00378. Elife. 2013. PMID: 23539368 Free PMC article. - CSF proteomics identifies early changes in autosomal dominant Alzheimer's disease.
Shen Y, Timsina J, Heo G, Beric A, Ali M, Wang C, Yang C, Wang Y, Western D, Liu M, Gorijala P, Budde J, Do A, Liu H, Gordon B, Llibre-Guerra JJ, Joseph-Mathurin N, Perrin RJ, Maschi D, Wyss-Coray T, Pastor P, Renton AE, Surace EI, Johnson ECB, Levey AI, Alvarez I, Levin J, Ringman JM, Allegri RF, Seyfried N, Day GS, Wu Q, Fernández MV, Tarawneh R, McDade E, Morris JC, Bateman RJ, Goate A; Dominantly Inherited Alzheimer Network; Ibanez L, Sung YJ, Cruchaga C. Shen Y, et al. Cell. 2024 Oct 31;187(22):6309-6326.e15. doi: 10.1016/j.cell.2024.08.049. Epub 2024 Sep 26. Cell. 2024. PMID: 39332414 Free PMC article. - Alcohol Activates Scabrous-Notch to Influence Associated Memories.
Petruccelli E, Feyder M, Ledru N, Jaques Y, Anderson E, Kaun KR. Petruccelli E, et al. Neuron. 2018 Dec 5;100(5):1209-1223.e4. doi: 10.1016/j.neuron.2018.10.005. Epub 2018 Oct 25. Neuron. 2018. PMID: 30482693 Free PMC article. - An Invasive Method for the Activation of the Mouse Dentate Gyrus by High-frequency Stimulation.
Zhao Z, Wu H. Zhao Z, et al. J Vis Exp. 2018 Jun 2;(136):57857. doi: 10.3791/57857. J Vis Exp. 2018. PMID: 29912203 Free PMC article.
References
- Bevins RA, Besheer J. Object recognition in rats and mice: a one-trial non-matching-to-sample learning task to study ‘recognition memory’. Nat Protoc. 2006;1:1306–1311. - PubMed
- Blackstone CD, Moss SJ, Martin LJ, Levey AI, Price DL, Huganir RL. Biochemical characterization and localization of a non-N-methyl-D-aspartate glutamate receptor in rat brain. J Neurochem. 1992;58:1118–1126. - PubMed
- Costa RM, Honjo T, Silva AJ. Learning and memory deficits in Notch mutant mice. Curr Biol. 2003;13:1348–1354. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous