A randomised trial of sheathed versus standard forceps for obtaining uncontaminated biopsy specimens of microbiota from the terminal ileum - PubMed (original) (raw)
Randomized Controlled Trial
A randomised trial of sheathed versus standard forceps for obtaining uncontaminated biopsy specimens of microbiota from the terminal ileum
Maneesh Dave et al. Gut. 2011 Aug.
Abstract
Background: The study of intestinal microbiota has been revolutionised by the use of molecular methods, including terminal restriction fragment length polymorphism (T-RFLP) analysis. Microbiota studies of Crohn's disease patients have examined samples from stool or from the neoterminal ileum with a standard biopsy forceps, which could be contaminated by colonic bacteria when the forceps passes through the colonoscope channel.
Objective: To determine whether sheathed biopsy forceps are able to obtain terminal ileal microbiota samples with less colonic bacterial contamination compared with unsheathed (standard) biopsy forceps.
Design: Prospective randomised single-centre study.
Patients and methods: Four (paired) biopsy specimens were obtained from adjacent locations in the terminal ileum using the sheathed and standard forceps of 27 consecutive subjects undergoing colonoscopy and the microbiota were characterised using T-RFLP. The Bray-Curtis similarity index between samples (sheathed vs unsheathed forceps) was calculated within patients and significant differences were tested for across all patients.
Results: There was not a significant difference in the microbial diversity of samples obtained using sheathed versus unsheathed forceps. The difference in microbial diversity between patients was much greater than the variability within patients by proximal versus distal site or by forceps type.
Limitations: T-RFLP is based on PCR amplification, so it is not always sensitive to rare bacterial species.
Conclusion: Standard unsheathed forceps appear to be sufficient for microbiota sample collection from the terminal ileum.
Conflict of interest statement
Disclosure:
The authors have no potential conflicts of interest or competing interests to disclose.
Figures
Figure 1
Bacterial Community Diversity Dendrogram. This neighbour-joining dendrogram was based on all pairwise comparisons between samples taken with sheathed (n=2) and unsheathed (n=2) forceps from 27 patients. Each sample is represented by a branch in the tree and branch lengths (the length of the line that connects two tips) represent the similarity among bacterial communities in these samples (the shorter the length, the more similar the communities).
Figure 2
Representative terminal restriction fragment length polymorphism (T-RFLP) chromatograms from unsheathed versus sheathed forceps. Representative T-RFLP profiles of unsheathed (A) compared to sheathed (B) biopsy samples from three patients. T-RFLP fragments (in base pairs) are represented as individual peaks. The peak height (relative fluorescence) corresponds to the relative abundance of each T-RFLP fragment. The most abundant individual fragments are labeled with respect to fragment size (in base pair) and relative percentage (decimal).
Figure 3
Microbial diversity measured by Bray-Curtis index (BCI). The mean difference in BCI between samples (pairwise analysis within individual patients) is presented. Error bars represent the SE of the mean. No significant differences by location or forceps type were found.
Figure 4
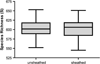
Terminal restriction fragment (TRF) richness (S). The middle line in the box represents the mean TRF richness. There is overlap between the TRF richness of unsheathed and sheathed forceps with the mean TRF richness of unsheathed forceps being only slightly greater than that of sheathed forceps (t-test, p=0.803).
Figure 5
Terminal restriction fragment (TRF) diversity as measured by the Shannon-Wiener diversity index (H’). The middle line represents the mean Shannon-Wiener diversity index for each group. The Shannon-Wiener diversity index for each individual patient is represented by a filled square (sheathed) or filled circle (unsheathed). The H’ overlaps between unsheathed and sheathed forceps with the mean H’ of unsheathed forceps being only slightly more than that of sheathed forceps. This is not statistically significant (t-test, p=0.316).
Similar articles
- Depth-dependent differences in community structure of the human colonic microbiota in health.
Lavelle A, Lennon G, Docherty N, Balfe A, Mulcahy HE, Doherty G, O Donoghue D, Hyland JM, Shanahan F, Sheahan K, Coffey JC, Winter DC, O Connell PR. Lavelle A, et al. PLoS One. 2013 Nov 6;8(11):e78835. doi: 10.1371/journal.pone.0078835. eCollection 2013. PLoS One. 2013. PMID: 24223167 Free PMC article. - Molecular analysis of jejunal, ileal, caecal and recto-sigmoidal human colonic microbiota using 16S rRNA gene libraries and terminal restriction fragment length polymorphism.
Hayashi H, Takahashi R, Nishi T, Sakamoto M, Benno Y. Hayashi H, et al. J Med Microbiol. 2005 Nov;54(Pt 11):1093-1101. doi: 10.1099/jmm.0.45935-0. J Med Microbiol. 2005. PMID: 16192442 - Polymerase chain reaction amplification of bacterial 16S rRNA genes from cold-cup biopsy forceps.
Keay S, Zhang CO, Baldwin BR, Alexander RB, Warren JW. Keay S, et al. J Urol. 1998 Dec;160(6 Pt 1):2229-31. doi: 10.1097/00005392-199812010-00091. J Urol. 1998. PMID: 9817375 - Influence of endoscopic biopsy forceps characteristics on tissue specimens: results of a prospective randomized study.
Woods KL, Anand BS, Cole RA, Osato MS, Genta RM, Malaty H, Gurer IE, Rossi DD. Woods KL, et al. Gastrointest Endosc. 1999 Feb;49(2):177-83. doi: 10.1016/s0016-5107(99)70483-9. Gastrointest Endosc. 1999. PMID: 9925695 Clinical Trial. - Contamination of single-use biopsy forceps: a prospective in vitro analysis.
Kinney TP, Kozarek RA, Raltz S, Attia F. Kinney TP, et al. Gastrointest Endosc. 2002 Aug;56(2):209-12. doi: 10.1016/s0016-5107(02)70179-x. Gastrointest Endosc. 2002. PMID: 12145598
Cited by
- Recent Advancements in Intestinal Microbiota Analyses: A Review for Non-Microbiologists.
Feng XW, Ding WP, Xiong LY, Guo L, Sun JM, Xiao P. Feng XW, et al. Curr Med Sci. 2018 Dec;38(6):949-961. doi: 10.1007/s11596-018-1969-z. Epub 2018 Dec 7. Curr Med Sci. 2018. PMID: 30536055 Review. - Microbiota of the Oropharynx and Endoscope Compared to the Esophagus.
Okereke IC, Miller AL, Hamilton CF, Booth AL, Reep GL, Andersen CL, Reynolds ST, Pyles RB. Okereke IC, et al. Sci Rep. 2019 Jul 15;9(1):10201. doi: 10.1038/s41598-019-46747-y. Sci Rep. 2019. PMID: 31308485 Free PMC article. - Spatial distribution of live gut microbiota and bile acid metabolism in various parts of human large intestine.
Chinda D, Takada T, Mikami T, Shimizu K, Oana K, Arai T, Akitaya K, Sakuraba H, Katto M, Nagara Y, Makino H, Fujii D, Oishi K, Fukuda S. Chinda D, et al. Sci Rep. 2022 Mar 4;12(1):3593. doi: 10.1038/s41598-022-07594-6. Sci Rep. 2022. PMID: 35246580 Free PMC article. - Isolation and Cultivation of Human Gut Microorganisms: A Review.
Wan X, Yang Q, Wang X, Bai Y, Liu Z. Wan X, et al. Microorganisms. 2023 Apr 20;11(4):1080. doi: 10.3390/microorganisms11041080. Microorganisms. 2023. PMID: 37110502 Free PMC article. Review. - Depth-dependent differences in community structure of the human colonic microbiota in health.
Lavelle A, Lennon G, Docherty N, Balfe A, Mulcahy HE, Doherty G, O Donoghue D, Hyland JM, Shanahan F, Sheahan K, Coffey JC, Winter DC, O Connell PR. Lavelle A, et al. PLoS One. 2013 Nov 6;8(11):e78835. doi: 10.1371/journal.pone.0078835. eCollection 2013. PLoS One. 2013. PMID: 24223167 Free PMC article.
References
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006 Feb 24;124(4):837–848. - PubMed
- Rutgeerts P, Van Assche G, Vermeire S, D'Haens G, Baert F, Noman M, et al. Ornidazole for prophylaxis of postoperative Crohn's disease recurrence: a randomized, double-blind, placebo-controlled trial. Gastroenterology. 2005 Apr;128(4):856–861. - PubMed
- Rutgeerts P, Hiele M, Geboes K, Peeters M, Penninckx F, Aerts R, et al. Controlled trial of metronidazole treatment for prevention of Crohn's recurrence after ileal resection. Gastroenterology. 1995 Jun;108(6):1617–1621. - PubMed
- Hayashi H, Sakamoto M, Benno Y. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol Immunol. 2002;46(8):535–548. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical