Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins - PubMed (original) (raw)
. 2011 Mar 15;108 Suppl 1(Suppl 1):4599-606.
doi: 10.1073/pnas.1000071108. Epub 2011 Feb 11.
Catherine A Lozupone, Federico E Rey, Meng Wu, Janaki L Guruge, Aneesha Narra, Jonathan Goodfellow, Jesse R Zaneveld, Daniel T McDonald, Julia A Goodrich, Andrew C Heath, Rob Knight, Jeffrey I Gordon
Affiliations
- PMID: 21317366
- PMCID: PMC3063581
- DOI: 10.1073/pnas.1000071108
Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins
Elizabeth E Hansen et al. Proc Natl Acad Sci U S A. 2011.
Abstract
The human gut microbiota harbors three main groups of H(2)-consuming microbes: methanogens including the dominant archaeon, Methanobrevibacter smithii, a polyphyletic group of acetogens, and sulfate-reducing bacteria. Defining their roles in the gut is important for understanding how hydrogen metabolism affects the efficiency of fermentation of dietary components. We quantified methanogens in fecal samples from 40 healthy adult female monozygotic (MZ) and 28 dizygotic (DZ) twin pairs, analyzed bacterial 16S rRNA datasets generated from their fecal samples to identify taxa that co-occur with methanogens, sequenced the genomes of 20 M. smithii strains isolated from families of MZ and DZ twins, and performed RNA-Seq of a subset of strains to identify their responses to varied formate concentrations. The concordance rate for methanogen carriage was significantly higher for MZ versus DZ twin pairs. Co-occurrence analysis revealed 22 bacterial species-level taxa positively correlated with methanogens: all but two were members of the Clostridiales, with several being, or related to, known hydrogen-producing and -consuming bacteria. The M. smithii pan-genome contains 987 genes conserved in all strains, and 1,860 variably represented genes. Strains from MZ and DZ twin pairs had a similar degree of shared genes and SNPs, and were significantly more similar than strains isolated from mothers or members of other families. The 101 adhesin-like proteins (ALPs) in the pan-genome (45 ± 6 per strain) exhibit strain-specific differences in expression and responsiveness to formate. We hypothesize that M. smithii strains use their different repertoires of ALPs to create diversity in their metabolic niches, by allowing them to establish syntrophic relationships with bacterial partners with differing metabolic capabilities and patterns of co-occurrence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
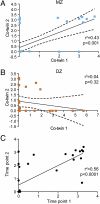
Correlation of methanogen levels in the fecal microbiota of MZ and DZ co-twins. The presence and levels of fecal methanogens were defined by qPCR assay that targeted the mcrA gene in samples obtained from MZ twin pairs (A) (n = 40) and DZ twin pairs (B) (n = 28). Dashed lines represent 95% confidence intervals for linear regression. (C) Correlation between mcrA levels in fecal samples collected at two time points per individual (2-mo interval between sampling). All axes in A_–_C are log10 (genome equivalents per ng total DNA +1).
Fig. 2.
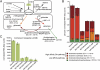
Normalized RNA-Seq reads assigned to the gene encoding an ammonium transporter (AmtB) and ECs involved in ammonia assimilation. (A) Overview of the two pathways of M. smithii for assimilating ammonia: The energy-dependent glutamine synthetase-glutamate synthase pathway has high affinity for ammonia (red arrow); an ATP-independent pathway has lower affinity (orange). (B) Strain-specific differences in the relative expression of components of the high affinity Gln pathway and the energy-independent low affinity pathway for ammonia assimilation. Mean values ± SEM are plotted. Colors represent components of the two pathways shown in A; color codes are coordinated between A and B. (C) Strain-specific differences in levels of expression of amtB. P < 0.0001 by one-way ANOVA.
Fig. 3.
Differential expression of M. smithii adhesin-like proteins (ALPs). Members of selected ALP OGUs with strain-specific differences in their expression profiles (A) and strain-specific, as well as OGU-associated, differences in their sensitivity to levels of formate during midlog phase growth (B). OGUs 112, 412, 827, and 208 exhibit strain-specific differences in their expression irrespective of formate concentration (one-way ANOVA, P < 0.0001), whereas OGUs 226, 287, 18, 133, and 37 contain at least one representative that is significantly regulated by formate concentration. Mean values ± SEM are plotted (n = 6 replicates per condition). * indicates a ≥2-fold difference, PPDE ≥ 0.97 (
Dataset S1, Table S7
).
Similar articles
- Mutual Exclusion of Methanobrevibacter Species in the Human Gut Microbiota Facilitates Directed Cultivation of a Candidatus Methanobrevibacter Intestini Representative.
Low A, Lee JKY, Gounot JS, Ravikrishnan A, Ding Y, Saw WY, Tan LWL, Moong DKN, Teo YY, Nagarajan N, Seedorf H. Low A, et al. Microbiol Spectr. 2022 Aug 31;10(4):e0084922. doi: 10.1128/spectrum.00849-22. Epub 2022 Jun 14. Microbiol Spectr. 2022. PMID: 35699469 Free PMC article. - Syntrophy via Interspecies H2 Transfer between Christensenella and Methanobrevibacter Underlies Their Global Cooccurrence in the Human Gut.
Ruaud A, Esquivel-Elizondo S, de la Cuesta-Zuluaga J, Waters JL, Angenent LT, Youngblut ND, Ley RE. Ruaud A, et al. mBio. 2020 Feb 4;11(1):e03235-19. doi: 10.1128/mBio.03235-19. mBio. 2020. PMID: 32019803 Free PMC article. - Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut.
Samuel BS, Hansen EE, Manchester JK, Coutinho PM, Henrissat B, Fulton R, Latreille P, Kim K, Wilson RK, Gordon JI. Samuel BS, et al. Proc Natl Acad Sci U S A. 2007 Jun 19;104(25):10643-8. doi: 10.1073/pnas.0704189104. Epub 2007 Jun 11. Proc Natl Acad Sci U S A. 2007. PMID: 17563350 Free PMC article. - Methanobrevibacter smithii cell variants in human physiology and pathology: A review.
Malat I, Drancourt M, Grine G. Malat I, et al. Heliyon. 2024 Sep 6;10(18):e36742. doi: 10.1016/j.heliyon.2024.e36742. eCollection 2024 Sep 30. Heliyon. 2024. PMID: 39347381 Free PMC article. Review. - Twin resemblance in somatotype and comparisons with other twin studies.
Song TM, Perusse L, Malina RM, Bouchard C. Song TM, et al. Hum Biol. 1994 Jun;66(3):453-64. Hum Biol. 1994. PMID: 8026815 Review.
Cited by
- Characteristics of the Gut Bacterial Composition in People of Different Nationalities and Religions.
Syromyatnikov M, Nesterova E, Gladkikh M, Smirnova Y, Gryaznova M, Popov V. Syromyatnikov M, et al. Microorganisms. 2022 Sep 18;10(9):1866. doi: 10.3390/microorganisms10091866. Microorganisms. 2022. PMID: 36144468 Free PMC article. Review. - Functional diversity within the simple gut microbiota of the honey bee.
Engel P, Martinson VG, Moran NA. Engel P, et al. Proc Natl Acad Sci U S A. 2012 Jul 3;109(27):11002-7. doi: 10.1073/pnas.1202970109. Epub 2012 Jun 18. Proc Natl Acad Sci U S A. 2012. PMID: 22711827 Free PMC article. - Vitamin B12 modulates the transcriptome of the skin microbiota in acne pathogenesis.
Kang D, Shi B, Erfe MC, Craft N, Li H. Kang D, et al. Sci Transl Med. 2015 Jun 24;7(293):293ra103. doi: 10.1126/scitranslmed.aab2009. Sci Transl Med. 2015. PMID: 26109103 Free PMC article. - The Archaeome's Role in Colorectal Cancer: Unveiling the DPANN Group and Investigating Archaeal Functional Signatures.
Mathlouthi NEH, Belguith I, Yengui M, Oumarou Hama H, Lagier JC, Ammar Keskes L, Grine G, Gdoura R. Mathlouthi NEH, et al. Microorganisms. 2023 Nov 10;11(11):2742. doi: 10.3390/microorganisms11112742. Microorganisms. 2023. PMID: 38004753 Free PMC article. - Genetic Determinants of the Gut Microbiome in UK Twins.
Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, Spector TD, Bell JT, Clark AG, Ley RE. Goodrich JK, et al. Cell Host Microbe. 2016 May 11;19(5):731-43. doi: 10.1016/j.chom.2016.04.017. Cell Host Microbe. 2016. PMID: 27173935 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- AA09022/AA/NIAAA NIH HHS/United States
- K05 AA017688/AA/NIAAA NIH HHS/United States
- P01 DK078669/DK/NIDDK NIH HHS/United States
- R01 DK030292/DK/NIDDK NIH HHS/United States
- K01 DK090285/DK/NIDDK NIH HHS/United States
- T32 GM07200-31/GM/NIGMS NIH HHS/United States
- DK70977/DK/NIDDK NIH HHS/United States
- T32 GM142607/GM/NIGMS NIH HHS/United States
- R01 AA017915/AA/NIAAA NIH HHS/United States
- R37 DK030292/DK/NIDDK NIH HHS/United States
- DK78669/DK/NIDDK NIH HHS/United States
- R01 DK070977/DK/NIDDK NIH HHS/United States
- T32 GM008759/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 AA009022/AA/NIAAA NIH HHS/United States
- T32 GM007200/GM/NIGMS NIH HHS/United States
- DK30292/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases