Cancer epigenetics: linking basic biology to clinical medicine - PubMed (original) (raw)
Review
Cancer epigenetics: linking basic biology to clinical medicine
Hsing-Chen Tsai et al. Cell Res. 2011 Mar.
Abstract
Cancer evolution at all stages is driven by both epigenetic abnormalities as well as genetic alterations. Dysregulation of epigenetic control events may lead to abnormal patterns of DNA methylation and chromatin configurations, both of which are critical contributors to the pathogenesis of cancer. These epigenetic abnormalities are set and maintained by multiple protein complexes and the interplay between their individual components including DNA methylation machinery, histone modifiers, particularly, polycomb (PcG) proteins, and chromatin remodeling proteins. Recent advances in genome-wide technology have revealed that the involvement of these dysregulated epigenetic components appears to be extensive. Moreover, there is a growing connection between epigenetic abnormalities in cancer and concepts concerning stem-like cell subpopulations as a driving force for cancer. Emerging data suggest that aspects of the epigenetic landscape inherent to normal embryonic and adult stem/progenitor cells may help foster, under the stress of chronic inflammation or accumulating reactive oxygen species, evolution of malignant subpopulations. Finally, understanding molecular mechanisms involved in initiation and maintenance of epigenetic abnormalities in all types of cancer has great potential for translational purposes. This is already evident for epigenetic biomarker development, and for pharmacological targeting aimed at reversing cancer-specific epigenetic alterations.
Figures
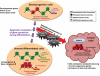
Figure 1
In normal stem/progenitor cells, the promoter regions of many CpG island-containing developmental genes are marked by both active (trimethylated histone H3 lysine 4; H3K4me3) and repressive marks (trimethylated histone H3 lysine 27; H3K27me3), termed “bivalent chromatin” by Bernstein et al. . This chromatin pattern holds these genes in a low, poised transcription state to prevent premature lineage commitment. When the stem/progenitor cells respond to environmental cues and start to differentiate, a shift of the balance between the active and repressive epigenetic marks takes place with corresponding changes in chromatin architecture, leading to the silencing of stemness genes and upregulation of lineage-specific genes. However, repeated environmental stress such as chronic inflammation or accumulating reactive oxygen species (ROS) may promote clonal expansion of cells with genetic or epigenetic abnormalities, which then contribute to tumor initiation and progression. During this course of oncogenesis, the repressive marks in the promoter regions of tumor suppressor genes may recruit DNA methylation machinery to impose abnormal CpG island methylation on these genes leading to permanent gene silencing. At the same time, these epigenetic abnormalities may also contribute to activation of stem cell pathways, such as the Wnt pathway, and bestow self-renewing properties on cancer cells.
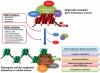
Figure 2
DNA methylation-mediated aberrant gene silencing in cancer involves transcriptional repressive complexes in the gene promoter region and interactions between DNA methylation machinery, chromatin modifiers (such as histone deacetylase, HDAC) and polycomb (PcG) proteins. Pharmacological inhibition of individual components in the repressive complex with DNMT inhibitors and HDAC inhibitors, either alone or in combination, may result in DNA demethylation and complex disintegration leading to reactivation of critical genes and reversal of genome-wide epigenetic alterations in cancer through resetting multiple cellular processes, including lineage commitment, immunomodulation, major cell signaling pathways, programmed cell death, and others. HAT: histone acetylase. Pol II: RNA polymerase II.
Similar articles
- Targeting cancer epigenetics: Linking basic biology to clinical medicine.
Shinjo K, Kondo Y. Shinjo K, et al. Adv Drug Deliv Rev. 2015 Dec 1;95:56-64. doi: 10.1016/j.addr.2015.10.006. Epub 2015 Oct 19. Adv Drug Deliv Rev. 2015. PMID: 26494398 Review. - Epigenetics in cancer: targeting chromatin modifications.
Ellis L, Atadja PW, Johnstone RW. Ellis L, et al. Mol Cancer Ther. 2009 Jun;8(6):1409-20. doi: 10.1158/1535-7163.MCT-08-0860. Epub 2009 Jun 9. Mol Cancer Ther. 2009. PMID: 19509247 Review. - Role of epigenetic modulation in cancer stem cell fate.
Deshmukh A, Binju M, Arfuso F, Newsholme P, Dharmarajan A. Deshmukh A, et al. Int J Biochem Cell Biol. 2017 Sep;90:9-16. doi: 10.1016/j.biocel.2017.07.003. Epub 2017 Jul 12. Int J Biochem Cell Biol. 2017. PMID: 28711634 Review. - Targeting Chromatin Remodeling for Cancer Therapy.
Kaur J, Daoud A, Eblen ST. Kaur J, et al. Curr Mol Pharmacol. 2019;12(3):215-229. doi: 10.2174/1874467212666190215112915. Curr Mol Pharmacol. 2019. PMID: 30767757 Free PMC article. Review. - Epigenetics in cancer.
Sharma S, Kelly TK, Jones PA. Sharma S, et al. Carcinogenesis. 2010 Jan;31(1):27-36. doi: 10.1093/carcin/bgp220. Epub 2009 Sep 13. Carcinogenesis. 2010. PMID: 19752007 Free PMC article. Review.
Cited by
- Integrated analysis of genome-wide DNA methylation and gene expression profiles identifies potential novel biomarkers of rectal cancer.
Wei J, Li G, Zhang J, Zhou Y, Dang S, Chen H, Wu Q, Liu M. Wei J, et al. Oncotarget. 2016 Sep 20;7(38):62547-62558. doi: 10.18632/oncotarget.11534. Oncotarget. 2016. PMID: 27566576 Free PMC article. - Epigenetic alterations involved in cancer stem cell reprogramming.
Muñoz P, Iliou MS, Esteller M. Muñoz P, et al. Mol Oncol. 2012 Dec;6(6):620-36. doi: 10.1016/j.molonc.2012.10.006. Epub 2012 Oct 26. Mol Oncol. 2012. PMID: 23141800 Free PMC article. Review. - Epigenetic alterations in patients with type 2 diabetes mellitus.
Karachanak-Yankova S, Dimova R, Nikolova D, Nesheva D, Koprinarova M, Maslyankov S, Tafradjiska R, Gateva P, Velizarova M, Hammoudeh Z, Stoynev N, Toncheva D, Tankova T, Dimova I. Karachanak-Yankova S, et al. Balkan J Med Genet. 2016 Jul 9;18(2):15-24. doi: 10.1515/bjmg-2015-0081. eCollection 2015 Dec 1. Balkan J Med Genet. 2016. PMID: 27785392 Free PMC article. - Epigenetics of Bladder Cancer: Where Biomarkers and Therapeutic Targets Meet.
Martinez VG, Munera-Maravilla E, Bernardini A, Rubio C, Suarez-Cabrera C, Segovia C, Lodewijk I, Dueñas M, Martínez-Fernández M, Paramio JM. Martinez VG, et al. Front Genet. 2019 Nov 18;10:1125. doi: 10.3389/fgene.2019.01125. eCollection 2019. Front Genet. 2019. PMID: 31850055 Free PMC article. Review. - The central role of EED in the orchestration of polycomb group complexes.
Cao Q, Wang X, Zhao M, Yang R, Malik R, Qiao Y, Poliakov A, Yocum AK, Li Y, Chen W, Cao X, Jiang X, Dahiya A, Harris C, Feng FY, Kalantry S, Qin ZS, Dhanasekaran SM, Chinnaiyan AM. Cao Q, et al. Nat Commun. 2014;5:3127. doi: 10.1038/ncomms4127. Nat Commun. 2014. PMID: 24457600 Free PMC article.
References
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. - PubMed
- Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 28:1057–1068. - PubMed
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. - PubMed
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources