Cohesin: genomic insights into controlling gene transcription and development - PubMed (original) (raw)
Review
Cohesin: genomic insights into controlling gene transcription and development
Dale Dorsett. Curr Opin Genet Dev. 2011 Apr.
Abstract
Over the past decade it has emerged that the cohesin protein complex, which functions in sister chromatid cohesion, chromosome segregation, and DNA repair, also regulates gene expression and development. Even minor changes in cohesin activity alter several aspects of development. Genome-wide analysis indicates that cohesin directly regulates transcription of genes involved in cell proliferation, pluripotency, and differentiation through multiple mechanisms. These mechanisms are poorly understood, but involve both partial gene repression in concert with Polycomb group proteins, and facilitating long-range looping, both between enhancers and promoters, and between CTCF protein binding sites.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
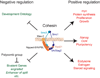
Figure 1
Cohesin regulates expression of genes that control differentiation, morphogenesis, proliferation, and pluripotency. The cohesin complex consists of the Smc1, Smc3, Rad21 and Stromalin (SA)/Stag2 subunits [3]. Cohesin is thought to bind by encircling DNA, and binding requires the Nipped-B/NIPBL – Mau-2 complex, and ATPase activity in the Smc1 and Smc3 head domains that interact with Rad21. Both the genes that increase or decrease in expression with reduced Nipped-B/NIPBL or cohesin activity are enriched for cohesin-binding genes, indicating that cohesin both represses and facilitates transcription. Developmental processes are the top gene ontology categories for genes that increase in expression when cohesin or Nipped-B/NIPBL activity is reduced in Drosophila cells derived from central nervous system [15*], and mouse embryonic stem cells [28*]. The majority of the genes that show the largest increases in expression are bivalent, with both histone H3 lysine 4 and lysine 27 trimethylation, indicating that they are also partially repressed by Polycomb group silencing proteins. Cohesin directly promotes expression of genes in the ecdysone steroid hormone signaling pathway in salivary glands and the mushroom body γ neuron [13,14,24*] and the estrogen pathway in human breast cancer cells [27*]. Nipped-B/NIPBL and cohesin also directly facilitate expression of myc genes, which promote protein synthesis and cell proliferation in all species examined [12*,15*,26*,28*,30,31,34*], and pluripotency genes in mouse embryonic stem cells [28*].
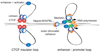
Figure 2
Cohesin facilitates DNA looping. The diagram shows two sister chromatids. On the left is a hypothetical model for how cohesin could support intrachromosomal looping between two CTCF binding sites, which occurs at several loci in mammalian cells [–39*]. In this example, the loop functions as an insulator, and sequesters a transcriptional enhancer, preventing it from activating flanking genes. The Nipped-B/NIPBL cohesin-loading factor is not present at CTCF binding sites [28*], suggesting that cohesin binds differently than at sites of sister chromatid cohesion. The right shows a model for how cohesin could stabilize a loop between an enhancer and promoter, facilitating transcriptional activation, such as occurs at the Nanog gene in mouse embryonic stem cells. The Mediator coactivator complex and Nipped-B/NIPBL are present [28*], and cohesin might function intrachromosomally in a manner similar to the way it mediates cohesion between sister chromatids.
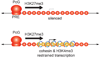
Figure 3
Cohesin acts in concert with Polycomb group (PcG) silencing proteins to restrain gene expression. In Drosophila cells, most PcG targeted genes are fully silenced and marked by histone H3 lysine 27 trimethylation (H3K27me3). They do not bind Nipped-B/NIPBL or cohesin [23*]. In rare cases, PcG targeted genes also bind cohesin, and all are expressed at low to moderate levels [15*]. All such genes encode transcription factors that control development. Nearly all genes targeted by both cohesin and PcG proteins are bivalent, having both the H3K27me3 mark, and the H3K4me3 modification associated with active genes. Reduction of either cohesin or Polycomb proteins strongly increases transcription of genes targeted simultaneously by cohesin and PcG proteins. The majority of genes that increase the most in expression with cohesin knockdown in mouse embryonic cells are also bivalent [28*,44], indicating that cohesin also contributes to repression of many PcG-targeted genes in mammalian cells.
Similar articles
- Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus.
Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Hadjur S, et al. Nature. 2009 Jul 16;460(7253):410-3. doi: 10.1038/nature08079. Epub 2009 May 20. Nature. 2009. PMID: 19458616 Free PMC article. - The Many Roles of Cohesin in Drosophila Gene Transcription.
Dorsett D. Dorsett D. Trends Genet. 2019 Jul;35(7):542-551. doi: 10.1016/j.tig.2019.04.002. Epub 2019 May 23. Trends Genet. 2019. PMID: 31130395 Free PMC article. Review. - Cohesin mediates transcriptional insulation by CCCTC-binding factor.
Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM. Wendt KS, et al. Nature. 2008 Feb 14;451(7180):796-801. doi: 10.1038/nature06634. Epub 2008 Jan 30. Nature. 2008. PMID: 18235444 - Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl.
Busslinger GA, Stocsits RR, van der Lelij P, Axelsson E, Tedeschi A, Galjart N, Peters JM. Busslinger GA, et al. Nature. 2017 Apr 27;544(7651):503-507. doi: 10.1038/nature22063. Epub 2017 Apr 19. Nature. 2017. PMID: 28424523 Free PMC article. - How cohesin and CTCF cooperate in regulating gene expression.
Wendt KS, Peters JM. Wendt KS, et al. Chromosome Res. 2009;17(2):201-14. doi: 10.1007/s10577-008-9017-7. Epub 2009 Mar 24. Chromosome Res. 2009. PMID: 19308701 Review.
Cited by
- Cohesins repress Kaposi's sarcoma-associated herpesvirus immediate early gene transcription during latency.
Chen HS, Wikramasinghe P, Showe L, Lieberman PM. Chen HS, et al. J Virol. 2012 Sep;86(17):9454-64. doi: 10.1128/JVI.00787-12. Epub 2012 Jun 27. J Virol. 2012. PMID: 22740398 Free PMC article. - Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres.
Remeseiro S, Cuadrado A, Carretero M, Martínez P, Drosopoulos WC, Cañamero M, Schildkraut CL, Blasco MA, Losada A. Remeseiro S, et al. EMBO J. 2012 May 2;31(9):2076-89. doi: 10.1038/emboj.2012.11. Epub 2012 Mar 13. EMBO J. 2012. PMID: 22415365 Free PMC article. - Cohesin and polycomb proteins functionally interact to control transcription at silenced and active genes.
Schaaf CA, Misulovin Z, Gause M, Koenig A, Gohara DW, Watson A, Dorsett D. Schaaf CA, et al. PLoS Genet. 2013 Jun;9(6):e1003560. doi: 10.1371/journal.pgen.1003560. Epub 2013 Jun 20. PLoS Genet. 2013. PMID: 23818863 Free PMC article. - Eukaryotic enhancers: common features, regulation, and participation in diseases.
Erokhin M, Vassetzky Y, Georgiev P, Chetverina D. Erokhin M, et al. Cell Mol Life Sci. 2015 Jun;72(12):2361-75. doi: 10.1007/s00018-015-1871-9. Epub 2015 Feb 26. Cell Mol Life Sci. 2015. PMID: 25715743 Free PMC article. Review. - Crosstalk between chromatin structure, cohesin activity and transcription.
Maya-Miles D, Andújar E, Pérez-Alegre M, Murillo-Pineda M, Barrientos-Moreno M, Cabello-Lobato MJ, Gómez-Marín E, Morillo-Huesca M, Prado F. Maya-Miles D, et al. Epigenetics Chromatin. 2019 Jul 22;12(1):47. doi: 10.1186/s13072-019-0293-6. Epigenetics Chromatin. 2019. PMID: 31331360 Free PMC article.
References
- Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–322. - PubMed
- Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. - PubMed
- Skibbens RV. Buck the establishment: reinventing sister chromatid cohesion. Trends Cell Biol. 2010;20:507–513. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous