HDAC3 and HDAC7 have opposite effects on osteoclast differentiation - PubMed (original) (raw)
HDAC3 and HDAC7 have opposite effects on osteoclast differentiation
Lan Pham et al. J Biol Chem. 2011.
Abstract
Histone deacetylases (HDACs) are negative regulators of transcription. Endochondral bone formation including chondrocyte and osteoblast maturation is regulated by HDACs. Very little is known about the role HDACs play in osteoclast differentiation. It has been previously reported that HDAC inhibitors, trichostatin A and sodium butyrate, suppress osteoclast differentiation through multiple mechanisms. In this study, we report that suppression of HDAC3 expression similar to HDAC inhibitors inhibits osteoclast differentiation, whereas osteoclasts suppressed for HDAC7 expression had accelerated differentiation when compared with control cells. Mitf, a transcription factor, is necessary for osteoclast differentiation. We demonstrate that Mitf and HDAC7 interact in RAW 264 cells and osteoclasts. The transcriptional activity of Mitf is repressed by HDAC7. Lastly, we show that either the amino or the carboxyl terminus of HDAC7 is sufficient for transcriptional repression and that the repression of HDAC7 is insensitive to trichostatin A, indicating that HDAC7 represses Mitf at least in part by deacetylation-independent mechanism.
Figures
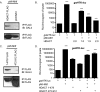
FIGURE 1.
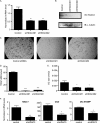
Suppression of HDAC3 inhibits osteoclast differentiation. A, real-time RT-PCR of bone marrow. **, p ≤ 0.007 versus control shRNA. B, Western blot (IB) of RAW 264.7 lysates showing expression of HDAC3. C, TRAP staining of osteoclast cultures. D and E, histomorphometric analysis of TRAP-stained osteoclasts (OC). MNCs, multinucleated cells. ***, p ≤ 0.0001 and *, p ≤ 0.05 versus control shRNA. F, expression profile of Nfat-c1, Ctsk, and DC-STAMP. *, p ≤ 0.05 and **, p ≤ 0.005 versus control shRNA following infection with control shRNA or HDAC3 shRNA lentiviral vectors.
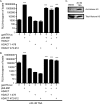
FIGURE 2.
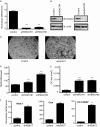
Accelerated osteoclast differentiation in HDAC7-suppressed osteoclasts. A, real-time RT-PCR of bone marrow. ***, p ≤ 0.0001 versus control shRNA. B, Western blot (IB) of osteoclast lysates showing expression of HDAC7. C, TRAP staining of osteoclast cultures infected with control or HDAC7 shRNA-expressing lentiviruses. D and E, histomorphometric analysis of TRAP-stained osteoclasts (OC). MNCs, multinucleated cells. ***, p ≤ 0.0001 versus control shRNA. F, expression profile of Nfat-c1, Ctsk, and DC-STAMP. **, p ≤ 0.001 versus control shRNA following infection with control shRNA or HDAC7 shRNA lentiviral vectors.
FIGURE 3.
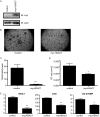
Overexpression of Myc-HDAC7 inhibits osteoclast differentiation. A, Western blot (IB) of osteoclast lysates. B, TRAP-stained images. C and D, histomorphometric analysis of TRAP-stained osteoclasts (OC). MNCs, multinucleated cells. ***, p ≤ 0.005 and *, p ≤ 0.05. E, expression profile of Nfat-c1, Ctsk, and DC-STAMP of osteoclast infected with control or Myc-HDAC7-expressing lentivirus. **, p ≤ 0.005 and *, p ≤ 0.05 versus control-infected cells.
FIGURE 4.
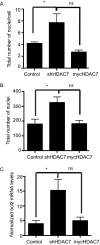
HDAC7 expression inhibits cell-cell fusion. Day 5 osteoclast cultures were stained with DAPI and Vybrant cell-labeling dye, and histomorphometry was performed to assess the total number of nuclei per multinucleated cell (A) and the total number of nuclei (B). *, p < 0.01 versus control-infected cells, ns = not significant when compared with control-infected cells. C, expression profile of bcl2 in osteoclasts infected with control, shHDAC7-, or Myc-HDAC7-overexpressing lentivirus. *, p < 0.01 versus control-infected cells, ns = not significant when compared with control-infected cells.
FIGURE 5.
HDAC7 interacts with Mitf. A and B, representative Western blots (IB) of Mitf immunoprecipitates (IP) from RAW 264.7 c4 cells (A) and primary mouse osteoclasts (B) immunoblotted against HDAC7 (Abcam). Osteoclasts or RAW 264.7 c4 cells were stimulated with M-CSF (10 ng/ml) for 2 days.
FIGURE 6.
Amino terminus of Mitf is sufficient for HDAC7 repression. A, 293T cells were transfected with FLAG-HDAC7 and pMI-Mitf. Lysates were immunoprecipitated (IP) with FLAG antibody and immunoblotted (IB) with an antibody that recognizes the Gal4 DNA binding domain. B, NIH 3T3 cells were transiently transfected with 5× Gal4-TK-luciferase reporter construct and/or pMI-Mitf and full-length HDAC7. Reporter activity is presented as relative luciferase units (RLU), and the results of three experiments each performed in duplicate are presented. ***, p ≤ 0.0001 _versus gal4-TK_-luc or _gal4-TK_-luc+Mitf. C, 293T cells were transfected with FLAG-HDAC7 1–478 or 472–912 and pMI-Mitf. Lysates were immunoprecipitated with FLAG antibody and immunoblotted with an antibody that recognizes the Gal4 DNA binding domain. D, NIH 3T3 cells were transiently transfected with 5× Gal4-TK-luciferase reporter construct and/or pMI-Mitf and either HDAC7 1–478 or HDAC7 472–912. Reporter activity is presented as relative luciferase units, and the results of three experiments each performed in duplicate are presented. ***, p ≤ 0.0001 _versus gal4-TK_-luc or _gal4-TK_-luc+Mitf.
FIGURE 7.
HDAC7 represses Mitf in a deacetylation-independent mechanism. A, NIH 3T3 cells were co-transfected with Gal4TK-luc, pMI-Mitf, and/or FLAG-HDAC7 1–428 or 422–912. Cells were treated with DMSO. Reporter activity is presented as relative luciferase units (RLU), and the results of three experiments each performed in duplicate are presented. B, repression of Mitf by HDAC7 is insensitive to TSA. Transfections were performed as described in A, and the cells were treated with 20 n
m
TSA for 16–20 h before harvest. ***, p ≤ 0.0001 _versus gal4-TK_-luc or _gal4-TK_-luc+Mitf. C, increased levels of acetylated histone H3 after TSA treatment. NIH 3T3 cells were treated with TSA in parallel to the experiment shown in B. Protein extracts were subjected to SDS-PAGE and immunoblotted against total and acetylated histone H3.
Similar articles
- Regulation of Osteoclast Differentiation and Skeletal Maintenance by Histone Deacetylases.
Faulkner B, Astleford K, Mansky KC. Faulkner B, et al. Molecules. 2019 Apr 6;24(7):1355. doi: 10.3390/molecules24071355. Molecules. 2019. PMID: 30959867 Free PMC article. Review. - Deletion of histone deacetylase 7 in osteoclasts decreases bone mass in mice by interactions with MITF.
Stemig M, Astelford K, Emery A, Cho JJ, Allen B, Huang TH, Gopalakrishnan R, Mansky KC, Jensen ED. Stemig M, et al. PLoS One. 2015 Apr 15;10(4):e0123843. doi: 10.1371/journal.pone.0123843. eCollection 2015. PLoS One. 2015. PMID: 25875108 Free PMC article. - Histone deacetylase 7 associates with Runx2 and represses its activity during osteoblast maturation in a deacetylation-independent manner.
Jensen ED, Schroeder TM, Bailey J, Gopalakrishnan R, Westendorf JJ. Jensen ED, et al. J Bone Miner Res. 2008 Mar;23(3):361-72. doi: 10.1359/jbmr.071104. J Bone Miner Res. 2008. PMID: 17997710 Free PMC article. - miRNA-340 inhibits osteoclast differentiation via repression of MITF.
Zhao H, Zhang J, Shao H, Liu J, Jin M, Chen J, Huang Y. Zhao H, et al. Biosci Rep. 2017 Jul 20;37(4):BSR20170302. doi: 10.1042/BSR20170302. Print 2017 Aug 31. Biosci Rep. 2017. PMID: 28607030 Free PMC article. - Mitf and Tfe3: members of a b-HLH-ZIP transcription factor family essential for osteoclast development and function.
Hershey CL, Fisher DE. Hershey CL, et al. Bone. 2004 Apr;34(4):689-96. doi: 10.1016/j.bone.2003.08.014. Bone. 2004. PMID: 15050900 Review.
Cited by
- Protein kinase D promotes in vitro osteoclast differentiation and fusion.
Mansky KC, Jensen ED, Davidova J, Yamamoto M, Gopalakrishnan R. Mansky KC, et al. J Biol Chem. 2013 Apr 5;288(14):9826-9834. doi: 10.1074/jbc.M112.444133. Epub 2013 Feb 21. J Biol Chem. 2013. PMID: 23430742 Free PMC article. - Regulation of Osteoclast Differentiation and Skeletal Maintenance by Histone Deacetylases.
Faulkner B, Astleford K, Mansky KC. Faulkner B, et al. Molecules. 2019 Apr 6;24(7):1355. doi: 10.3390/molecules24071355. Molecules. 2019. PMID: 30959867 Free PMC article. Review. - A transcriptional network underlies susceptibility to kidney disease progression.
Laouari D, Burtin M, Phelep A, Bienaime F, Noel LH, Lee DC, Legendre C, Friedlander G, Pontoglio M, Terzi F. Laouari D, et al. EMBO Mol Med. 2012 Aug;4(8):825-39. doi: 10.1002/emmm.201101127. Epub 2012 Jun 18. EMBO Mol Med. 2012. PMID: 22711280 Free PMC article. - The underestimated role of the microphthalmia-associated transcription factor (MiTF) in normal and pathological haematopoiesis.
Oppezzo A, Rosselli F. Oppezzo A, et al. Cell Biosci. 2021 Jan 13;11(1):18. doi: 10.1186/s13578-021-00529-0. Cell Biosci. 2021. PMID: 33441180 Free PMC article. Review. - Chromatin modifiers and histone modifications in bone formation, regeneration, and therapeutic intervention for bone-related disease.
Gordon JAR, Stein JL, Westendorf JJ, van Wijnen AJ. Gordon JAR, et al. Bone. 2015 Dec;81:739-745. doi: 10.1016/j.bone.2015.03.011. Epub 2015 Mar 31. Bone. 2015. PMID: 25836763 Free PMC article. Review.
References
- Sengupta N., Seto E. (2004) J. Cell. Biochem. 93, 57–67 - PubMed
- Gregoretti I. V., Lee Y. M., Goodson H. V. (2004) J. Mol. Biol. 338, 17–31 - PubMed
- Drummond D. C., Noble C. O., Kirpotin D. B., Guo Z., Scott G. K., Benz C. C. (2005) Annu. Rev. Pharmacol. Toxicol. 45, 495–528 - PubMed
- Luo R. X., Postigo A. A., Dean D. C. (1998) Cell 92, 463–473 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AR056642/AR/NIAMS NIH HHS/United States
- R03 DE020117/DE/NIDCR NIH HHS/United States
- R03 DE020117-01A1/DE/NIDCR NIH HHS/United States
- T32 DE007288/DE/NIDCR NIH HHS/United States
LinkOut - more resources
Full Text Sources