Isolation and characterisation of KP34--a novel φKMV-like bacteriophage for Klebsiella pneumoniae - PubMed (original) (raw)
Isolation and characterisation of KP34--a novel φKMV-like bacteriophage for Klebsiella pneumoniae
Zuzanna Drulis-Kawa et al. Appl Microbiol Biotechnol. 2011 May.
Abstract
Bacteriophage KP34 is a novel virus belonging to the subfamily Autographivirinae lytic for extended-spectrum β-lactamase-producing Klebsiella pneumoniae strains. Its biological features, morphology, susceptibility to chemical and physical agents, burst size, host specificity and activity spectrum were determined. As a potential antibacterial agent used in therapy, KP34 molecular features including genome sequence and protein composition were examined. Phylogenetic analyses and clustering of KP34 phage genome sequences revealed its clear relationships with "phiKMV-like viruses". Simultaneously, whole-genome analyses permitted clustering and classification of all phages, with completely sequenced genomes, belonging to the Podoviridae.
Figures
Fig. 1
Electron micrograph of phage KP34 negatively stained with uranyl acetate. The bar indicates 100 nm
Fig. 2
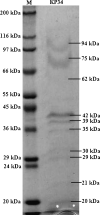
SDS-PAGE analysis of purified KP34 virus particles with Sigma-Aldrich wide-range molecular weight markers in the left lane
Fig. 3
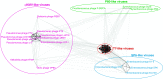
Graphic layout made in CLANS software for 36 Autographivirinae genome sequences using BLASTN searches. Analysed sequences are represented by vertices connected by edges reflecting attractive forces proportional to the negative logarithm of the HSP’s P value. The greyness intensity of the connections is proportional to these forces. The not shown phages names forming compact clusters are fully listed in Table S2 in electronic supplementary material
Fig. 4
Phyml tree of tail tubular proteins A (a) and tail tubular proteins B (b). Sequence from Klebsiella phage KP34 indicated in bold font. Sequences from phages are placed in the grey rectangle. Numbers at nodes, in the shown order, correspond to the minimum of support values calculated in Phyml by _χ_2 and Shimodaira–Hasegawa-like procedure (χ 2-SH); support values resulted from bootstrap analysis in Phyml (PH) and posterior probabilities estimated in Phylobayes (PB). Values of the posterior probabilities and bootstrap percentages lower or equal to 0.50 and 50%, respectively, were omitted or indicated by a dash. GenBank identifiers (gi) of sequences are shown in parenthesis
Fig. 4
Phyml tree of tail tubular proteins A (a) and tail tubular proteins B (b). Sequence from Klebsiella phage KP34 indicated in bold font. Sequences from phages are placed in the grey rectangle. Numbers at nodes, in the shown order, correspond to the minimum of support values calculated in Phyml by _χ_2 and Shimodaira–Hasegawa-like procedure (χ 2-SH); support values resulted from bootstrap analysis in Phyml (PH) and posterior probabilities estimated in Phylobayes (PB). Values of the posterior probabilities and bootstrap percentages lower or equal to 0.50 and 50%, respectively, were omitted or indicated by a dash. GenBank identifiers (gi) of sequences are shown in parenthesis
Fig. 5
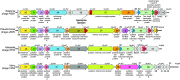
Comparison of 3′-end genomic sequence in four phages classified to “φKMV-like” viruses. ORFs whose products show significant sequence similarity are presented in the same colour. ORFs in grey have their products annotated as hypothetical protein
Similar articles
- Biological characteristics and genome analysis of a novel phage vB_KpnP_IME279 infecting Klebsiella pneumoniae.
Zhang R, Zhao F, Wang J, Pei G, Fan H, Zhangxiang L, Mi Z, Shi T, Liu H, Tong Y. Zhang R, et al. Folia Microbiol (Praha). 2020 Dec;65(6):925-936. doi: 10.1007/s12223-020-00775-8. Epub 2020 Oct 16. Folia Microbiol (Praha). 2020. PMID: 33064268 - Characterising the biology of novel lytic bacteriophages infecting multidrug resistant Klebsiella pneumoniae.
Kęsik-Szeloch A, Drulis-Kawa Z, Weber-Dąbrowska B, Kassner J, Majkowska-Skrobek G, Augustyniak D, Lusiak-Szelachowska M, Zaczek M, Górski A, Kropinski AM. Kęsik-Szeloch A, et al. Virol J. 2013 Mar 28;10:100. doi: 10.1186/1743-422X-10-100. Virol J. 2013. PMID: 23537199 Free PMC article. - Discovery of Three Toxic Proteins of Klebsiella Phage fHe-Kpn01.
Spruit CM, Wicklund A, Wan X, Skurnik M, Pajunen MI. Spruit CM, et al. Viruses. 2020 May 15;12(5):544. doi: 10.3390/v12050544. Viruses. 2020. PMID: 32429141 Free PMC article. - Comparative genome analysis of novel Podoviruses lytic for hypermucoviscous Klebsiella pneumoniae of K1, K2, and K57 capsular types.
Solovieva EV, Myakinina VP, Kislichkina AA, Krasilnikova VM, Verevkin VV, Mochalov VV, Lev AI, Fursova NK, Volozhantsev NV. Solovieva EV, et al. Virus Res. 2018 Jan 2;243:10-18. doi: 10.1016/j.virusres.2017.09.026. Epub 2017 Oct 5. Virus Res. 2018. PMID: 28988127 - How to introduce a new bacteriophage on the block: a short guide to phage classification.
Valencia-Toxqui G, Ramsey J. Valencia-Toxqui G, et al. J Virol. 2024 Oct 22;98(10):e0182123. doi: 10.1128/jvi.01821-23. Epub 2024 Sep 12. J Virol. 2024. PMID: 39264154 Review.
Cited by
- In Vitro Activity, Stability and Molecular Characterization of Eight Potent Bacteriophages Infecting Carbapenem-Resistant Klebsiella pneumoniae.
Baqer AA, Fang K, Mohd-Assaad N, Adnan SNA, Md Nor NS. Baqer AA, et al. Viruses. 2022 Dec 30;15(1):117. doi: 10.3390/v15010117. Viruses. 2022. PMID: 36680156 Free PMC article. - Genome Sequence of KP-Rio/2015, a Novel Klebsiella pneumoniae (Podoviridae) Phage.
Meira GL, Campos FS, Albuquerque JP, Cabral MC, Fracalanzza SE, Campos RM, Vermelho AB, Ferreira DF. Meira GL, et al. Genome Announc. 2016 Dec 22;4(6):e01298-16. doi: 10.1128/genomeA.01298-16. Genome Announc. 2016. PMID: 28007847 Free PMC article. - Luminescent Phage-Based Detection of Klebsiella pneumoniae: From Engineering to Diagnostics.
Zelcbuch L, Yitzhaki E, Nissan O, Gidron E, Buchshtab N, Kario E, Kredo-Russo S, Zak NB, Bassan M. Zelcbuch L, et al. Pharmaceuticals (Basel). 2021 Apr 9;14(4):347. doi: 10.3390/ph14040347. Pharmaceuticals (Basel). 2021. PMID: 33918942 Free PMC article. - Complete genomic sequence of bacteriophage P23: a novel Vibrio phage isolated from the Yellow Sea, China.
Liu Y, Zhao L, Wang M, Wang Q, Zhang X, Han Y, Wang M, Jiang T, Shao H, Jiang Y, McMinn A. Liu Y, et al. Virus Genes. 2019 Dec;55(6):834-842. doi: 10.1007/s11262-019-01699-3. Epub 2019 Aug 16. Virus Genes. 2019. PMID: 31420829 - Isolation, characterization, and antibacterial activity of lytic bacteriophage against methicillin-resistant Staphylococcus aureus causing bedsore and diabetic wounds.
Rezaei Z, Elikaei A, Barzi SM, Shafiei M. Rezaei Z, et al. Iran J Microbiol. 2022 Oct;14(5):712-720. doi: 10.18502/ijm.v14i5.10967. Iran J Microbiol. 2022. PMID: 36531807 Free PMC article.
References
- Ackermann HW, DuBow MS. Viruses of prokaryotes. Boca Raton: CRC; 1987. pp. 143–172.
- Adams M. Bacteriophages. New York: Interscience; 1959. pp. 137–159.
- Bogovazova GG, Voroshilova NN, Bondarenko VM. The efficacy of Klebsiella pneumoniae bacteriophage in the therapy of experimental Klebsiella infection. Zh Mikrobiol Epidemiol Immunobiol. 1991;4:5–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials