Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity - PubMed (original) (raw)
doi: 10.1038/nm.2313. Epub 2011 Feb 20.
Jin Chen, Alejandra Petrilli, Geraldine Liot, Eva Klinglmayr, Yue Zhou, Patrick Poquiz, Jonathan Tjong, Mahmoud A Pouladi, Michael R Hayden, Eliezer Masliah, Mark Ellisman, Isabelle Rouiller, Robert Schwarzenbacher, Blaise Bossy, Guy Perkins, Ella Bossy-Wetzel
Affiliations
- PMID: 21336284
- PMCID: PMC3051025
- DOI: 10.1038/nm.2313
Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity
Wenjun Song et al. Nat Med. 2011 Mar.
Abstract
Huntington's disease is an inherited and incurable neurodegenerative disorder caused by an abnormal polyglutamine (polyQ) expansion in huntingtin (encoded by HTT). PolyQ length determines disease onset and severity, with a longer expansion causing earlier onset. The mechanisms of mutant huntingtin-mediated neurotoxicity remain unclear; however, mitochondrial dysfunction is a key event in Huntington's disease pathogenesis. Here we tested whether mutant huntingtin impairs the mitochondrial fission-fusion balance and thereby causes neuronal injury. We show that mutant huntingtin triggers mitochondrial fragmentation in rat neurons and fibroblasts of individuals with Huntington's disease in vitro and in a mouse model of Huntington's disease in vivo before the presence of neurological deficits and huntingtin aggregates. Mutant huntingtin abnormally interacts with the mitochondrial fission GTPase dynamin-related protein-1 (DRP1) in mice and humans with Huntington's disease, which, in turn, stimulates its enzymatic activity. Mutant huntingtin-mediated mitochondrial fragmentation, defects in anterograde and retrograde mitochondrial transport and neuronal cell death are all rescued by reducing DRP1 GTPase activity with the dominant-negative DRP1 K38A mutant. Thus, DRP1 might represent a new therapeutic target to combat neurodegeneration in Huntington's disease.
Figures
Figure 1
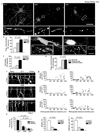
Mutant HTT triggers mitochondrial fragmentation, decrease in anterograde and retrograde transport, and neuronal cell death, which depends on polyQ length. (a) Fluorescence micrographs and ×6 zoom of boxed regions of neurons expressing HTT exon1-Q17, -Q46, or -Q97 and DsRed2-Mito. Scale bar, 50 µm. (b) Mitochondrial fragmentation of neurons expressing HTT exon1-Q17, -Q46, or -Q97 and DsRed2-Mito. (c) Cell death of neurons expressing HTT exon1-Q17, -Q46, or -Q97. (d) Fluorescence micrographs and ×3 zoom of boxed regions of MitoTracker Red stained human fibroblasts from an unaffected (left) or an adult onset HD individual (right). Scale bar, 25 µm. (e) Mitochondrial fragmentation in fibroblasts from an unaffected or HD individuals. (f) Kymographs of mitochondrial transport in neurons expressing HTT exon1-Q17, -Q46, or -Q97 and DsRed2-Mito. See also Movies S1, S2, S3. (g) Scatter plots of mitochondrial velocity in retrograde or anterograde direction as a function of distance traveled in 5 minutes in neurons (n = 10) expressing HTT exon1-Q17, -Q46, or -Q97 and DsRed2-Mito. (h) Anterograde and retrograde movement, motility, and mean velocity of mitochondria in the same neurons analyzed in (g). Data are mean ± s.e.m. of triplicate samples of representative experiments. Results are representative of three or more independent experiments. Statistics: one-way ANOVA test.
Figure 2
Mutant HTT increases the number of small mitochondria and cristae, but decreases cristae surface area and volume in the striatum of six-month old YAC128 mice. (a) EM of control (top) and YAC128 (bottom) brain showing an elongated neuronal mitochondrion (arrowhead) and several short mitochondria (arrowheads), respectively. Scale bar, 500 nm. (b) Percentage of mitochondria of short length (0–1,000 nm) and long length (1,000–5,000 nm). (c) EM of a control mitochondrion (arrowhead) nearly 4 µm long. (d) Surface-rendered volume showing the normal structure of its 84 cristae. (e) EM of a mitochondrion fissioning into three parts (arrowheads). (f) A slice of the volume shows the separation of the three mitochondrial bodies (arrows). (g) Top view showing all 223 cristae. There are no cristae in the constriction (arrowhead) between the left and middle bodies, but a few cristae appear in the constriction between the middle and right bodies (arrow). Side view of the outer membrane showing the width of the constrictions. Side view demonstrating cristae fragmentation by how few cristae extend from top to bottom of the volume. (h) Number of cristae per mitochondrial cross-sectional area. (i, j) Cristae surface area and volume. Data are mean ± s.e.m. Statistics: Student’s _t_-test, P < 0.05*, P < 0.01**.
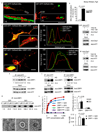
Figure 3
Mutant HTT interacts with DRP1 in HD mice and human and alters DRP1 structure and function. (a) Fluorescence micrographs of neurons expressing HTT exon1 and DsRed2-Mito. Scale bar, 50 µm. (b) Co-localization of mitochondria and HTT in neurons co-expressing HTT exon1 and DsRed2-Mito. (c) Fluorescence micrographs with line scan of co-localization of HTT, DRP1, and mitochondria in neurons co-expressing DsRed2-Mito, _DRP1_-YFP, and HTT exon1. Scale bar, 10 µm. (d) Co-immune precipitations of brain mitochondrial lysates from 1.5 or 2 month old YAC18 and YAC128 mice followed with HTT or Drp1 antibodies. The intensities of the signals are presented as arbitrary units (A.U.) and are normalized to input signals. (e) Co-immune precipitations of human lymphoblast lysates from unaffected or HD individuals with HTT antibodies. (f) Co-immune precipitations of human postmortem brain tissue lysates from unaffected or HD individuals with DRP1 or HTT antibodies. (g) Co-immune precipitations of bacterial DRP1 protein and bacterial HTT exon1-Q20-GST or -Q53-GST protein with DRP1 antibodies. (h) Steady-state kinetics of DRP1 GTPase activity (left), bar graph of GTPase activity at 0.05 mM GTP and apparent Michaelis-Menten constant (Km) (right) in the presence of wild-type or mutant HTT exon1 protein and MOM liposomes. Data are mean ± s.d. from three independent measurements. Statistics: Student’s _t_-test. (i) Negative stain EM images of baculovirus DRP1 in the absence of nucleotides (left), the presence of GTP (center), and the presence of GTP and HTT exon1-Q53 -GST protein (right). Scale bars, 10 nm.
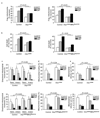
Figure 4
Restoring mitochondrial fusion with DRP1K38A or in combination with MFN2RasG12V rescues neurons from neuritic trafficking defects and cell death. (a) Mitochondrial fragmentation of neurons expressing either HTT exon1-Q17, -Q46, or -Q97 alone or in combination with either _DRP1_K38A alone, or _DRP1_K38A and _MFN2_RasG12V. (b) Cell death of neurons expressing either mutant HTT exon1-Q17, -Q46, or -Q97 and DsRed2-Mito alone or in combination with either _DRP1_K38A alone or _DRP1_K38A and _MFN2_RasG12V. (c) Anterograde and retrograde movement of mitochondria in neurons expressing either HTT exon1-Q17, -Q46, or -Q97 alone or in combination with _DRP1_K38A alone or _DRP1_K38A and _MFN2_RasG12V. (d) Motility of mitochondria in neurons expressing either HTT exon1-Q17, -Q46, or -Q97 alone or in combination with either _DRP1_K38A alone or both _DRP1_K38A and _MFN2_RasG12V. (e) Mean velocity of mitochondria in neurons expressing either HTT exon1-Q17, -Q46, or -Q97 alone or in combination with _DRP1_K38A alone or _DRP1_K38A and _MFN2_RasG12V. Results are representative of three or more independent experiments. Statistics: Two-way ANOVA with post-hoc test.
Comment in
- Hugging tight in Huntington's.
Johri A, Chaturvedi RK, Beal MF. Johri A, et al. Nat Med. 2011 Mar;17(3):245-6. doi: 10.1038/nm0311-245. Nat Med. 2011. PMID: 21383715 No abstract available.
Similar articles
- Mutant huntingtin's interaction with mitochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington's disease.
Shirendeb UP, Calkins MJ, Manczak M, Anekonda V, Dufour B, McBride JL, Mao P, Reddy PH. Shirendeb UP, et al. Hum Mol Genet. 2012 Jan 15;21(2):406-20. doi: 10.1093/hmg/ddr475. Epub 2011 Oct 13. Hum Mol Genet. 2012. PMID: 21997870 Free PMC article. - S-nitrosylation of dynamin-related protein 1 mediates mutant huntingtin-induced mitochondrial fragmentation and neuronal injury in Huntington's disease.
Haun F, Nakamura T, Shiu AD, Cho DH, Tsunemi T, Holland EA, La Spada AR, Lipton SA. Haun F, et al. Antioxid Redox Signal. 2013 Oct 10;19(11):1173-84. doi: 10.1089/ars.2012.4928. Epub 2013 Jun 20. Antioxid Redox Signal. 2013. PMID: 23641925 Free PMC article. - Effects of overexpression of huntingtin proteins on mitochondrial integrity.
Wang H, Lim PJ, Karbowski M, Monteiro MJ. Wang H, et al. Hum Mol Genet. 2009 Feb 15;18(4):737-52. doi: 10.1093/hmg/ddn404. Epub 2008 Nov 27. Hum Mol Genet. 2009. PMID: 19039036 Free PMC article. - Increased mitochondrial fission and neuronal dysfunction in Huntington's disease: implications for molecular inhibitors of excessive mitochondrial fission.
Reddy PH. Reddy PH. Drug Discov Today. 2014 Jul;19(7):951-5. doi: 10.1016/j.drudis.2014.03.020. Epub 2014 Mar 28. Drug Discov Today. 2014. PMID: 24681059 Free PMC article. Review. - Mutant huntingtin, abnormal mitochondrial dynamics, defective axonal transport of mitochondria, and selective synaptic degeneration in Huntington's disease.
Reddy PH, Shirendeb UP. Reddy PH, et al. Biochim Biophys Acta. 2012 Feb;1822(2):101-10. doi: 10.1016/j.bbadis.2011.10.016. Epub 2011 Nov 4. Biochim Biophys Acta. 2012. PMID: 22080977 Free PMC article. Review.
Cited by
- Signaling Pathways Concerning Mitochondrial Dysfunction: Implications in Neurodegeneration and Possible Molecular Targets.
Sharma Y, Gupta JK, Babu MA, Singh S, Sindhu RK. Sharma Y, et al. J Mol Neurosci. 2024 Oct 28;74(4):101. doi: 10.1007/s12031-024-02269-5. J Mol Neurosci. 2024. PMID: 39466510 Review. - Mitochondrial Dynamics: A Key Role in Neurodegeneration and a Potential Target for Neurodegenerative Disease.
Yang D, Ying J, Wang X, Zhao T, Yoon S, Fang Y, Zheng Q, Liu X, Yu W, Hua F. Yang D, et al. Front Neurosci. 2021 Apr 12;15:654785. doi: 10.3389/fnins.2021.654785. eCollection 2021. Front Neurosci. 2021. PMID: 33912006 Free PMC article. Review. - Metformin Protects Cells from Mutant Huntingtin Toxicity Through Activation of AMPK and Modulation of Mitochondrial Dynamics.
Jin J, Gu H, Anders NM, Ren T, Jiang M, Tao M, Peng Q, Rudek MA, Duan W. Jin J, et al. Neuromolecular Med. 2016 Dec;18(4):581-592. doi: 10.1007/s12017-016-8412-z. Epub 2016 May 25. Neuromolecular Med. 2016. PMID: 27225841 Free PMC article. - Antioxidants in Huntington's disease.
Johri A, Beal MF. Johri A, et al. Biochim Biophys Acta. 2012 May;1822(5):664-74. doi: 10.1016/j.bbadis.2011.11.014. Epub 2011 Nov 23. Biochim Biophys Acta. 2012. PMID: 22138129 Free PMC article. Review. - Mitochondrial and Redox Modifications in Huntington Disease Induced Pluripotent Stem Cells Rescued by CRISPR/Cas9 CAGs Targeting.
Lopes C, Tang Y, Anjo SI, Manadas B, Onofre I, de Almeida LP, Daley GQ, Schlaeger TM, Rego ACC. Lopes C, et al. Front Cell Dev Biol. 2020 Sep 22;8:576592. doi: 10.3389/fcell.2020.576592. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33072759 Free PMC article.
References
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS055193/NS/NINDS NIH HHS/United States
- R01NS055193/NS/NINDS NIH HHS/United States
- R01EY016164/EY/NEI NIH HHS/United States
- P41 RR004050-21/RR/NCRR NIH HHS/United States
- R01 NS047456-05/NS/NINDS NIH HHS/United States
- P41 RR004050-24/RR/NCRR NIH HHS/United States
- P41 RR004050/RR/NCRR NIH HHS/United States
- P41RR004050/RR/NCRR NIH HHS/United States
- R01 NS047456/NS/NINDS NIH HHS/United States
- R01 EY016164/EY/NEI NIH HHS/United States
- CAPMC/ CIHR/Canada
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous