Metabolic regulation by p53 - PubMed (original) (raw)
Review
Metabolic regulation by p53
Oliver D K Maddocks et al. J Mol Med (Berl). 2011 Mar.
Erratum in
- J Mol Med. 2011 May;89(5):531
Abstract
We are increasingly aware that cellular metabolism plays a vital role in diseases such as cancer, and that p53 is an important regulator of metabolic pathways. By transcriptional activation and other means, p53 is able to contribute to the regulation of glycolysis, oxidative phosphorylation, glutaminolysis, insulin sensitivity, nucleotide biosynthesis, mitochondrial integrity, fatty acid oxidation, antioxidant response, autophagy and mTOR signalling. The ability to positively and negatively regulate many of these pathways, combined with feedback signalling from these pathways to p53, demonstrates the reciprocal and flexible nature of the regulation, facilitating a diverse range of responses to metabolic stress. Intriguingly, metabolic stress triggers primarily an adaptive (rather than pro-apoptotic) p53 response, and p53 is emerging as an important regulator of metabolic homeostasis. A better understanding of how p53 coordinates metabolic adaptation will facilitate the identification of novel therapeutic targets and will also illuminate the wider role of p53 in human biology.
Figures
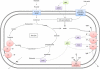
Fig. 1
p53 signalling in glucose metabolism. p53 can suppress the transcription of glucose transporters GLUT1 and GLUT4 (and via NFκB inhibits GLUT3) along with the insulin receptor to inhibit cellular glucose uptake. By transcriptional activation of TIGAR, p53 can suppress the rate of glycolysis and increase diversion of glycolytic intermediates into the PPP. p53 can also suppress glycolysis by promoting the degradation of phosphoglycerate mutase (PGM). By activating the transcription of hexokinase II (HK II) p53 can stimulate glycolysis. Malate dehydrogenase (MDH1) forms part of the malate/aspartate shuttle that links glycolysis to mitochondrial respiration; MDH1 binds to and modulates the activity of p53
Fig. 2
p53 and mitochondrial respiration. Basal p53 levels transcriptionally activate synthesis of cytochrome oxidase 2 (SCO2) and apoptosis-inducing factor (AIF), which support the function of mitochondrial respiratory chain complexes I, III & IV and acts directly on complex IV subunit 1. p53 transcriptionally activates glutaminase 2 (GLS2), which catalyses the conversion of glutamine to glutamate. p53 regulates FAO via transcriptional activation of guanidinoacetate aminotransferase (GAMT), and possibly by other mechanisms
Fig. 3
Signalling between p53 and the IGF-AKT-mTORC1 pathway. p53 is activated by metabolic stress signals via AMPK, ATM and mTORC1-S6K-MDM2 signalling, and modulates energy metabolism, cell cycle and autophagy. p53 can inhibit mTORC1 signalling via transcriptional activation of PTEN, IGF-BP3, Sestrin1 & Sestrin2 (SES1/2), TSC2 and AMPKβ. Direct interaction of p53 with LKB1 may directly lead to AMPK activation. Dotted lines indicate transcriptional regulation, unbroken lines indicate a direct protein level effect, arrowheads indicate an activating signal, blunt-ended lines indicate an inhibitory signal
References
- Kondoh H, Lleonart ME, Gil J, Wang J, Degan P, Peters G, Martinez D, Carnero A, Beach D. Glycolytic enzymes can modulate cellular life span. Cancer Res. 2005;65:177–185. - PubMed
- Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous