Functional analysis of KAP1 genomic recruitment - PubMed (original) (raw)
Functional analysis of KAP1 genomic recruitment
Sushma Iyengar et al. Mol Cell Biol. 2011 May.
Abstract
TRIM28 (KAP1) is upregulated in many cancers and has been implicated in both transcriptional activation and repression. Using chromatin immunoprecipitation and sequencing, we show that KAP1 binding sites fall into several categories, specifically, the 3' coding exons of zinc finger (ZNF) genes and promoter regions of ZNFs and other genes. The currently accepted model is that KAP1 is recruited to the genome via interaction of its N-terminal RBCC domain with KRAB ZNFs (KRAB domain containing ZNFs). To determine whether the interaction of KAP1 with KRAB ZNFs is the mechanism by which KAP1 is recruited to genomic binding sites, we analyzed stable cell lines that express tagged wild-type and mutant KAP1. Surprisingly, deletion of the RBCC domain abolished KAP1 binding to the 3' exons of ZNF genes but KAP1 binding to promoter regions was unaffected. Using KAP1 knockdown cells, we showed that the genes most responsive to KAP1 were not ZNF genes but instead were either indirect targets or had KAP1 bound 10 to 100 kb from the transcription start site. Therefore, our studies suggest that KAP1 plays a role distinct from transcriptional regulation at the majority of its strongest binding sites.
Figures
Fig. 1.
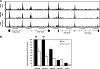
Description and expression of KAP1 mutants. (A) Illustration of endogenous KAP1 protein and the KAP1 mutants used in this study. Well-characterized protein interaction domains of KAP1 are indicated along with their interacting partners. All of the mutant constructs, as well as WT KAP1, were tagged with the FLAG peptide at the N terminus. (B) Western blot analysis of KAP1 mutant stable cell lines. Plasmids bearing the FLAG vector, FLAG-tagged WT KAP1, or FLAG-tagged mutant KAP1 constructs were stably transfected into HEK293 cells. Shown are Western analyses of nuclear extract from untransfected HEK293 cells probed with a KAP1 antibody (lane 1); nuclear extract from HEK293 cells harboring the empty FLAG vector, probed with the anti-FLAG antibody (lane 2); and nuclear extracts from HEK293 cells harboring various KAP1 constructs, probed with the anti-FLAG antibody (lanes 3 to 7). The positions of the molecular weight markers are shown on the left. Each lane was also probed with an antibody to actin as a loading control.
Fig. 2.
Comparison of FLAG-tagged and endogenous KAP1. (A) ChIP-seq binding patterns. Snapshots of ChIP-seq results for endogenous KAP1 in U2OS cells, endogenous KAP1 in HEK293 cells, and FLAG-tagged WT KAP1 in HEK293 cells for a region on chromosome 19. The number of sequenced tags is shown on the y axis, and the coordinates of several ZNF genes are shown on the x axis. (B) ChIP-PCR confirmation of KAP1 binding sites. ChIPs were performed with HEK293 cells stably expressing FLAG-tagged WT KAP1. Antibodies that recognize KAP1 or the FLAG tag were used. Five binding sites identified by ChIP-seq were confirmed by qPCR using independent ChIP samples. The fold enrichment of KAP1 and FLAG-tagged WT KAP1 (compared to 0.1% of the starting chromatin) is shown; results are from two independent replicates; the error bars indicate standard errors.
Fig. 3.
KAP1 binds to promoters and nonpromoter regions. (A) Five binding sites identified by ChIP-seq were selected from each of three different types of KAP1 targets, i.e., 3′ ends of ZNF genes, 5′ regions of ZNF genes, and 5′ regions of non-ZNF genes; 4 negative-control regions were also chosen. All regions were tested by ChIP-qPCR using an antibody against the FLAG peptide and HEK293 cells stably expressing FLAG-tagged WT KAP1. Fold enrichment for FLAG-tagged WT KAP1 (compared to 0.1% of the starting chromatin) is shown; results are from two independent replicates, and the error bars indicate standard errors. (B) Shown are ChIP-seq profiles for FLAG-tagged WT KAP1 for an example from each category of KAP1 binding sites. The number of sequenced tags is shown on the y axis, and the genomic coordinates are shown on the x axis. The arrow indicates the direction of transcription.
Fig. 4.
The SETDB1 histone methylation complex is recruited to specific subsets of KAP1 targets. ChIP-qPCR data for H3K9me3 (A), SetDB1 (B), H3K36me3 (C), and H3K4me3 (D) at the chosen binding sites (see Fig. 3) from HEK293 cells stably expressing FLAG-tagged WT KAP1. Fold enrichment (compared to 0.1% of the starting chromatin) is shown; results are from two independent replicates, and the error bars indicate standard errors.
Fig. 5.
Interaction with HP1 is required for efficient nuclear retention of KAP1. Immunofluorescence staining was performed with HEK293 cells stably expressing either WT or mutant KAP1 proteins using antibodies against the FLAG tag (green) or KAP1 (red); nuclei were stained with DAPI (blue).
Fig. 6.
Interaction with HP1 is required for efficient tethering of KAP1 to DNA. (A) ChIP-qPCR was performed using an antibody against the FLAG tag and HEK293 cells stably expressing FLAG-tagged M2-KAP1 (this KAP1 construct harbors two amino acid substitutions in the HP1 binding domain). Fold enrichment of FLAG-tagged M2-KAP1 (compared to 0.1% of the starting chromatin) is shown; results are from two independent replicates, and the error bars indicate standard errors. (B) Shown are ChIP-seq profiles for FLAG-tagged M2-KAP1 for an example from each category of KAP1 binding sites. The number of sequenced tags is shown on the y axis, and the genomic coordinates are shown on the x axis. The arrows indicate the direction of transcription. (C) Snapshot of ChIP-seq data for FLAG-tagged M2-KAP1, compared to WT KAP1, at a region on chromosome 19. The number of sequenced tags is shown on the y axis, and the genomic coordinates are shown on the x axis. The arrows indicate the direction of transcription.
Fig. 7.
The RBCC domain of KAP1 is required for recruitment to the 3′ ends of ZNF genes. (A) ChIP-qPCR was performed using an antibody against the FLAG tag and HEK293 cells stably expressing FLAG-tagged ΔRBCC-KAP1 (this KAP1 construct contains a deletion of the domain that interacts with KRAB ZNFs). Fold enrichment for FLAG-tagged ΔRBCC-KAP1 (compared to 0.1% of the starting chromatin) is shown; results are from two independent replicates, and the error bars indicate standard errors. (B) Shown are ChIP-seq profiles for an example from each category of KAP1 binding sites for FLAG-tagged ΔRBCC-KAP1. The number of sequenced tags is shown on the y axis, and the genomic coordinates are shown on the x axis. The arrows indicate the direction of transcription. (C) Snapshot of ChIP-seq data for FLAG-tagged ΔRBCC-KAP1, compared to WT KAP1, at a region on chromosome 19. The number of sequenced tags is shown on the y axis, and the genomic coordinates are shown on the x axis. The arrows indicate the direction of transcription.
Fig. 8.
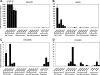
Functional annotation of WT KAP1 versus ΔRBCC-KAP1 binding sites. Gene ontology analyses (right panels) and location analyses (left panels) are shown for all FLAG-tagged WT KAP1 peaks (A), the top 1,000 FLAG-tagged WT KAP1 peaks (B), and all FLAG-tagged ΔRBCC-KAP1 (∼1,000 total) peaks (C). For the gene ontology analyses, the nearest gene to each binding site was identified and then the program DAVID (6, 13) was used to identify enriched categories; redundant categories were eliminated, and the top 10 categories based on their P value are shown. Ontology terms are shown on the y axis; P values for the significance of enrichment are graphed along the x axis. For the location analyses, the distance between the binding sites and the closest transcription start site (using the UCSC KnownGenes HG19) is shown. Distances were calculated from the center of the KAP1 binding site to the nearest transcription start site and binned in progressively larger intervals between 100 kb upstream and 100 kb downstream of the transcription start site.
Fig. 9.
The PHD and bromodomain of KAP1 are not required for recruitment to DNA. (A) ChIP-qPCR was performed using an antibody against the FLAG tag and HEK293 cells stably expressing FLAG-tagged ΔPB-KAP1 (this KAP1 construct contains a deletion of the C terminus of KAP1 that removes the PHD and bromodomain). Fold enrichment for FLAG-tagged ΔPB-KAP1 (compared to 0.1% of the starting chromatin) is shown; results are from two independent replicates, and the error bars indicate standard errors. (B) Shown are ChIP-seq profiles for an example from each category of KAP1 binding sites for FLAG-tagged ΔPB-KAP1. The number of sequenced tags is shown on the y axis, and the genomic coordinates are shown on the x axis. The arrows indicate the direction of transcription. (C) Snapshot of ChIP-seq data for FLAG-tagged ΔPB-KAP1, compared to WT KAP1, at a region on chromosome 19. The number of sequenced tags is shown on the y axis, and the genomic coordinates are shown on the x axis. The arrows indicate the direction of transcription.
Fig. 10.
ChIP-seq analysis of KAP1 lacking both the N- and C-terminal domains. (A) ChIP-qPCR was performed using an antibody against the FLAG tag and HEK293 cells stably expressing FLAG-tagged Δ(RBBB+PB)-KAP1 (this KAP1 construct contains a deletion of both the N and the C termini of KAP1). Fold enrichment for Δ(RBBB+PB)-KAP1 (compared to 0.1% of the starting chromatin) is shown; results are from two independent replicates, and the error bars indicate standard errors. (B) Shown are ChIP-seq profiles for an example from each category of KAP1 binding sites for Δ(RBBB+PB)-KAP1. The number of sequenced tags is shown on the y axis, and the genomic coordinates are shown on the x axis. The arrows indicate the direction of transcription. (C) Snapshot of ChIP-seq data for FLAG-tagged Δ(RBBB+PB)-KAP1, compared to WT KAP1, at a region on chromosome 19. The number of sequenced tags is shown on the y axis, and the genomic coordinates are shown on the x axis. The arrows indicate the direction of transcription.
Fig. 11.
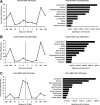
Analysis of KAP1 knockdown cells. (A) Shown is a Western blot analysis of KAP1 levels in control (NT2 GFP) and knockdown (NT2 K2 GFP) cells. (B) ChIP-PCR of a positive KAP1 binding site (ZNF180) and a negative-control region (EVX1) in control cells (NT2 GFP) and KAP1 knockdown cells (NT2 K2 GFP). RNA from control and KAP1 knockdown cells was analyzed on Illumina Sentrix Beadchip arrays. The fold changes in expression of the genes activated (C) or repressed (D) by KAP1 are shown. The top 100 mRNAs that responded to loss of KAP1 in the activated and repressed sets were further analyzed. First, the mRNAs were compared to the KAP1 target list to determine which were directly regulated by KAP1; 52 of the top 100 activated genes and 38 of the top 100 repressed genes were KAP1 targets. Then, a location analysis was performed on the 52 directly activated (E) and 38 directly repressed (F) KAP1 targets. The binding sites are plotted relative to the start site of transcription of the target gene.
Fig. 12.
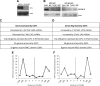
ChIP-seq patterns at genes regulated by KAP1. Shown are ChIP-seq patterns for several genes activated (A and B) or repressed (C and D) by KAP1. The binding of KAP1 to EFEMP1, EPAS1, ZSWIM5, and NRXN2 was also confirmed by ChIP-PCR (data not shown).
Similar articles
- Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs.
O'Geen H, Squazzo SL, Iyengar S, Blahnik K, Rinn JL, Chang HY, Green R, Farnham PJ. O'Geen H, et al. PLoS Genet. 2007 Jun;3(6):e89. doi: 10.1371/journal.pgen.0030089. Epub 2007 Apr 19. PLoS Genet. 2007. PMID: 17542650 Free PMC article. - ZNF274 recruits the histone methyltransferase SETDB1 to the 3' ends of ZNF genes.
Frietze S, O'Geen H, Blahnik KR, Jin VX, Farnham PJ. Frietze S, et al. PLoS One. 2010 Dec 8;5(12):e15082. doi: 10.1371/journal.pone.0015082. PLoS One. 2010. PMID: 21170338 Free PMC article. - ATRX binds to atypical chromatin domains at the 3' exons of zinc finger genes to preserve H3K9me3 enrichment.
Valle-García D, Qadeer ZA, McHugh DS, Ghiraldini FG, Chowdhury AH, Hasson D, Dyer MA, Recillas-Targa F, Bernstein E. Valle-García D, et al. Epigenetics. 2016 Jun 2;11(6):398-414. doi: 10.1080/15592294.2016.1169351. Epub 2016 Mar 30. Epigenetics. 2016. PMID: 27029610 Free PMC article. - KAP1/TRIM28: Transcriptional Activator and/or Repressor of Viral and Cellular Programs?
Randolph K, Hyder U, D'Orso I. Randolph K, et al. Front Cell Infect Microbiol. 2022 Feb 23;12:834636. doi: 10.3389/fcimb.2022.834636. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35281453 Free PMC article. Review. - KRAB zinc finger proteins.
Ecco G, Imbeault M, Trono D. Ecco G, et al. Development. 2017 Aug 1;144(15):2719-2729. doi: 10.1242/dev.132605. Development. 2017. PMID: 28765213 Free PMC article. Review.
Cited by
- Breaking through an epigenetic wall: re-activation of Oct4 by KRAB-containing designer zinc finger transcription factors.
Juárez-Moreno K, Erices R, Beltran AS, Stolzenburg S, Cuello-Fredes M, Owen GI, Qian H, Blancafort P. Juárez-Moreno K, et al. Epigenetics. 2013 Feb;8(2):164-76. doi: 10.4161/epi.23503. Epub 2013 Jan 11. Epigenetics. 2013. PMID: 23314702 Free PMC article. - KRAB-ZFP Transcriptional Regulators Acting as Oncogenes and Tumor Suppressors: An Overview.
Sobocińska J, Molenda S, Machnik M, Oleksiewicz U. Sobocińska J, et al. Int J Mol Sci. 2021 Feb 23;22(4):2212. doi: 10.3390/ijms22042212. Int J Mol Sci. 2021. PMID: 33672287 Free PMC article. Review. - The KRAB zinc finger protein RSL1 regulates sex- and tissue-specific promoter methylation and dynamic hormone-responsive chromatin configuration.
Krebs CJ, Schultz DC, Robins DM. Krebs CJ, et al. Mol Cell Biol. 2012 Sep;32(18):3732-42. doi: 10.1128/MCB.00615-12. Epub 2012 Jul 16. Mol Cell Biol. 2012. PMID: 22801370 Free PMC article. - Oncogenesis driven by the Ras/Raf pathway requires the SUMO E2 ligase Ubc9.
Yu B, Swatkoski S, Holly A, Lee LC, Giroux V, Lee CS, Hsu D, Smith JL, Yuen G, Yue J, Ann DK, Simpson RM, Creighton CJ, Figg WD, Gucek M, Luo J. Yu B, et al. Proc Natl Acad Sci U S A. 2015 Apr 7;112(14):E1724-33. doi: 10.1073/pnas.1415569112. Epub 2015 Mar 24. Proc Natl Acad Sci U S A. 2015. PMID: 25805818 Free PMC article. - TRIM28 mediates chromatin modifications at the TCRα enhancer and regulates the development of T and natural killer T cells.
Zhou XF, Yu J, Chang M, Zhang M, Zhou D, Cammas F, Sun SC. Zhou XF, et al. Proc Natl Acad Sci U S A. 2012 Dec 4;109(49):20083-8. doi: 10.1073/pnas.1214704109. Epub 2012 Nov 19. Proc Natl Acad Sci U S A. 2012. PMID: 23169648 Free PMC article.
References
- Barski A., et al. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823–837 - PubMed
- Dehal P., et al. 2001. Human chromosome 19 and related regions in mouse: conservative and lineage-specific evolution. Science 293:104–111 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54HG004558/HG/NHGRI NIH HHS/United States
- U54 HG004558/HG/NHGRI NIH HHS/United States
- P20 RR016440/RR/NCRR NIH HHS/United States
- CA45240/CA/NCI NIH HHS/United States
- R01 CA045240/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous