Lumenal interactions in nuclear pore complex assembly and stability - PubMed (original) (raw)
Lumenal interactions in nuclear pore complex assembly and stability
William T Yewdell et al. Mol Biol Cell. 2011.
Abstract
Nuclear pore complexes (NPCs) provide a gateway for the selective transport of macromolecules across the nuclear envelope (NE). Although we have a solid understanding of NPC composition and structure, we do not have a clear grasp of the mechanism of NPC assembly. Here, we demonstrate specific defects in nucleoporin distribution in strains lacking Heh1p and Heh2p-two conserved members of the LEM (Lap2, emerin, MAN1) family of integral inner nuclear membrane proteins. These effects on nucleoporin localization are likely of functional importance as we have defined specific genetic interaction networks between HEH1 and HEH2, and genes encoding nucleoporins in the membrane, inner, and outer ring complexes of the NPC. Interestingly, expression of a domain of Heh1p that resides in the NE lumen is sufficient to suppress both the nucleoporin mislocalization and growth defects in heh1Δpom34Δ strains. We further demonstrate a specific physical interaction between the Heh1p lumenal domain and the massive cadherin-like lumenal domain of the membrane nucleoporin Pom152p. These findings support a role for Heh1p in the assembly or stability of the NPC, potentially through the formation of a lumenal bridge with Pom152p.
Figures
FIGURE 1:
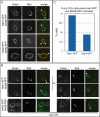
Specific defects in NPC distribution in heh1Δ and heh2Δ strains. Fluorescence micrographs (top panels) of deconvolved images of GFP-Nup49p, Mlp1-GFP, and Esc1-GFP in either heh1Δ, heh2Δ, or heh1Δheh2Δ strains. A merge between phase-contrast images and the fluorescent images are shown in bottom panels. To better visualize the cytoplasmic accumulation of GFP-Nup49 foci (arrows), a maximum intensity projection (MIP) is shown. Note that in heh2Δ cells there are few cytoplasmic GFP-Nup49p foci and the NPCs appear clustered at the NE.
FIGURE 2:
Heh proteins colocalize with a fraction of Nic96-RFP. The colocalization of endogenous levels of Heh1- and Heh2-GFP was determined with Nic96-RFP in WT (A) and nup133Δ strains where NPCs are clustered at the NE (B). To achieve a high spatial resolution, the cells were fixed and immobilized prior to imaging, and images were subsequently deconvolved. Arrows in merged images point to regions of overlap between the green and red channels. In (B), colocalization in nup133Δ cells was often only observed in one axial plane. The top two image series are identical cells separated by 0.2 μm in the z direction (Δz). (C) Quantitation of the percentage of nup133Δ cells with at least one region of colocalization of Heh1- or Heh2-GFP and Nic96-RFP.
FIGURE 3:
Affinity purification of Heh1p and Heh2p demonstrates specific interactions with nups (A and B). Magnetic beads with α-GFP antibodies were used to pull out either Heh1-GFP or Heh2-GFP (or no GFP; B) from cell lysates derived from strains expressing HEH1-GFP or HEH2-GFP and the indicated c-myc- or HA-tagged nups. After washing, α-GFP beads were eluted with SDS–PAGE sample buffer. Proteins were separated by SDS–PAGE and Western blotted with the indicated antibodies (left) and were detected by HRP-conjugated secondary antibodies and enhanced chemiluminescence. Equivalent amounts of cells were used for each experiment. Equivalent amounts of load (L) and unbound (UB) fractions are shown in left panels and represent ∼0.5% of the total extract. Right panels show proteins in bound fractions to either Heh1-GFP (B1) or Heh2-GFP (B2), and represent 20% of bound proteins. For each row, the right and left panels can be directly compared, as they are the same membrane with identical exposure times.
FIGURE 4:
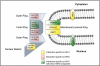
Schematic of HEH1 and HEH2 interactions with nup genes. Genes encoding nuclear basket, membrane, inner, and outer ring complexes are shown in the context of the approximate location of their gene products relative to the POM. (Gray circles with tails represent phospholipids.) Blue-gray polygons are physical representations of the indicated nup subcomplexes. Interactions are colored as described in the key. The schematic is not intended to reflect the stoichiometry of these subcomplexes in the NPC.
FIGURE 5:
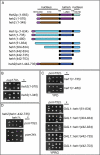
Lumenal and nuclear domains of Heh1p and Heh2p contribute specificity to the functional interactions with the inner and membrane ring nups. (A) Schematic of N- and C-terminal deletion constructs of Heh1p and Heh2p. Heh1p and Heh2p share a similar domain architecture and topology: a helix-extension helix HEH/LEM domain and NTD, two transmembrane domains (TM), a lumenal domain (LUMD), and another nuclear domain (MCHD). Numbers are amino acid residues. (B–D) Colonies from tetrad dissections (in rows) of genetic crosses between the indicated strains on YPD or YPRG (for expression of GAL1 alleles). In (B) and (C), the underlined colony expresses an heh allele in the indicated deletion strain. In (D), underlined colonies represent, in descending order: heh1Δpom152Δ and heh1Δpom34Δ. Dashed underlined colonies are the double knockout strains expressing the heh2(heh1–442-735) allele. Note that heh2(heh1–442-735) rescues the growth and viability of both heh1Δpom34Δ and heh1Δpom152Δ strains.
FIGURE 6:
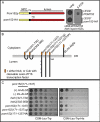
Heh1p and Pom152p lumenal domains specifically interact. (A) Schematic showing both full-length Pom152p with NPC, TM, and lumenal (lumen) domains, in addition to the product of an allele of POM152 with two tandem HA peptides (pom152-HA) inserted into the lumenal domain. As shown at right, this allele is unable to support growth of heh1Δpom152Δ strains. An heh1Δpom152Δ strain covered with a URA3/POM152 plasmid and transformed with either a LEU2/POM152 or LEU2/pom152-HA plasmid was plated onto YPD or 5-FOA (to force the loss of URA3/POM152). Images were taken after 2 d at 30°C. (B) Schematic of lumenal domain constructs and topology in ER (gray lines are monolayers) expressed as fusions to both the N (Nub/prey/2μm/TRP1) or C terminus (Cub/bait/CEN/LEU2) of ubiquitin. Cub is fused to a cleavable LexA-VP16 transcription factor released upon Cub-Nub interaction that promotes transcription of the HIS3 gene in the two-hybrid query strain (NMY32). Positive interactions are thus assessed as growth on CSM-Leu-Trp-His. AI-Alg5 and DL-Alg5 are positive (+) and negative (–) controls for bait topology and self-activation, respectively. Numbers are amino acids. Y’s are glycosylation sites. HA is two tandem HA peptides inserted into the Pom152p lumenal domain. (C) NMY32 was simultaneously transformed with plasmids expressing the bait/Cub-fusion of pom152(171–1337) and the indicated prey/Nub fusions. Transformants were spotted in 10-fold serial dilutions on CSM-Leu-Trp plates and CSM-Leu-Trp-His plates and imaged after 3 d at 30°C.
FIGURE 7:
The Heh1p lumenal domain can rescue nup mislocalization in heh1Δpom34Δ cells. (A) Maximum intensity projections of a deconvolved z-series showing the subcellular distribution of Nup82-GFP and Nup60-GFP in heh1Δpom34Δ cells. These images are merged with a phase-contrast image in right panels. Cytoplasmic nup foci are indicated by arrows. The mislocalization of Nup60-GFP is unique, and it appears in a reticular pattern (arrowheads). The mislocalization of Nup82-GFP can be rescued by the expression of the heh2(heh1–442-735) allele (+heh1–442-735). (B) Quantitation of the percentage of cells showing nup mislocalization in the indicated strains and rescue with the heh2(heh1–442-735) allele. (C) A time-lapse series (Δt is 30 min) showing the mislocalization of Nup60-GFP in heh1Δpom34Δ cells delayed in mitosis (see Supplemental Movie S1). Left and middle panels are maximum intensity projections of a z-series of images of Nup60-GFP and Nic96-RFP, respectively, and the right panels are a merge of green, red, and phase-contrast images. Cells marked as 1, 2, or 3 are all proceeding into mitosis. By 90 min, all three cells show a redistribution of both Nup60-GFP and Nic96-RFP, most strikingly in cell 3, where there is a mitotic catastrophe that results in a loss of NE organization.
Similar articles
- Heh2/Man1 may be an evolutionarily conserved sensor of NPC assembly state.
Borah S, Thaller DJ, Hakhverdyan Z, Rodriguez EC, Isenhour AW, Rout MP, King MC, Lusk CP. Borah S, et al. Mol Biol Cell. 2021 Jul 15;32(15):1359-1373. doi: 10.1091/mbc.E20-09-0584. Epub 2021 May 19. Mol Biol Cell. 2021. PMID: 34010011 Free PMC article. - Nuclear pore complex function in Saccharomyces cerevisiae is influenced by glycosylation of the transmembrane nucleoporin Pom152p.
Belanger KD, Gupta A, MacDonald KM, Ott CM, Hodge CA, Cole CM, Davis LI. Belanger KD, et al. Genetics. 2005 Nov;171(3):935-47. doi: 10.1534/genetics.104.036319. Epub 2005 Aug 22. Genetics. 2005. PMID: 16118201 Free PMC article. - Chm7 and Heh1 collaborate to link nuclear pore complex quality control with nuclear envelope sealing.
Webster BM, Thaller DJ, Jäger J, Ochmann SE, Borah S, Lusk CP. Webster BM, et al. EMBO J. 2016 Nov 15;35(22):2447-2467. doi: 10.15252/embj.201694574. Epub 2016 Oct 12. EMBO J. 2016. PMID: 27733427 Free PMC article. - Nuclear envelope insertion of spindle pole bodies and nuclear pore complexes.
Jaspersen SL, Ghosh S. Jaspersen SL, et al. Nucleus. 2012 May-Jun;3(3):226-36. doi: 10.4161/nucl.20148. Epub 2012 May 1. Nucleus. 2012. PMID: 22572959 Free PMC article. Review. - The Nuclear Pore Complex: Birth, Life, and Death of a Cellular Behemoth.
Dultz E, Wojtynek M, Medalia O, Onischenko E. Dultz E, et al. Cells. 2022 Apr 25;11(9):1456. doi: 10.3390/cells11091456. Cells. 2022. PMID: 35563762 Free PMC article. Review.
Cited by
- Quality control mechanisms that protect nuclear envelope identity and function.
Mannino PJ, Lusk CP. Mannino PJ, et al. J Cell Biol. 2022 Sep 5;221(9):e202205123. doi: 10.1083/jcb.202205123. Epub 2022 Aug 29. J Cell Biol. 2022. PMID: 36036741 Free PMC article. Review. - Fission yeast Lem2 and Man1 perform fundamental functions of the animal cell nuclear lamina.
Gonzalez Y, Saito A, Sazer S. Gonzalez Y, et al. Nucleus. 2012 Jan-Feb;3(1):60-76. doi: 10.4161/nucl.18824. Nucleus. 2012. PMID: 22540024 Free PMC article. - Trafficking to uncharted territory of the nuclear envelope.
Burns LT, Wente SR. Burns LT, et al. Curr Opin Cell Biol. 2012 Jun;24(3):341-9. doi: 10.1016/j.ceb.2012.01.009. Epub 2012 Feb 10. Curr Opin Cell Biol. 2012. PMID: 22326668 Free PMC article. Review. - Fantastic nuclear envelope herniations and where to find them.
Thaller DJ, Patrick Lusk C. Thaller DJ, et al. Biochem Soc Trans. 2018 Aug 20;46(4):877-889. doi: 10.1042/BST20170442. Epub 2018 Jul 19. Biochem Soc Trans. 2018. PMID: 30026368 Free PMC article. Review. - The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteasome.
Niepel M, Molloy KR, Williams R, Farr JC, Meinema AC, Vecchietti N, Cristea IM, Chait BT, Rout MP, Strambio-De-Castillia C. Niepel M, et al. Mol Biol Cell. 2013 Dec;24(24):3920-38. doi: 10.1091/mbc.E13-07-0412. Epub 2013 Oct 23. Mol Biol Cell. 2013. PMID: 24152732 Free PMC article.
References
- Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1997.
- Alber F, et al. Determining the architectures of macromolecular assemblies. Nature. 2007a;450:683–694. - PubMed
- Alber F, et al. The molecular architecture of the nuclear pore complex. Nature. 2007b;450:695–701. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous