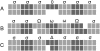RNAcode: robust discrimination of coding and noncoding regions in comparative sequence data - PubMed (original) (raw)
RNAcode: robust discrimination of coding and noncoding regions in comparative sequence data
Stefan Washietl et al. RNA. 2011 Apr.
Abstract
With the availability of genome-wide transcription data and massive comparative sequencing, the discrimination of coding from noncoding RNAs and the assessment of coding potential in evolutionarily conserved regions arose as a core analysis task. Here we present RNAcode, a program to detect coding regions in multiple sequence alignments that is optimized for emerging applications not covered by current protein gene-finding software. Our algorithm combines information from nucleotide substitution and gap patterns in a unified framework and also deals with real-life issues such as alignment and sequencing errors. It uses an explicit statistical model with no machine learning component and can therefore be applied "out of the box," without any training, to data from all domains of life. We describe the RNAcode method and apply it in combination with mass spectrometry experiments to predict and confirm seven novel short peptides in Escherichia coli and to analyze the coding potential of RNAs previously annotated as "noncoding." RNAcode is open source software and available for all major platforms at http://wash.github.com/rnacode.
Figures
FIGURE 1.
Overview of the RNAcode algorithm. First, a phylogenetic tree is estimated from the input alignment including a reference sequence (darker line) under a noncoding (neutral) nucleotide model. From this background model and a protein similarity matrix, a normalized substitution score is derived to evaluate observed mutations for evidence of negative selection. This substitution score and a gap scoring scheme are the basis for a dynamic programming (DP) algorithm to find local high-scoring coding segments. To estimate the statistical significance of these segments, a background score distribution is estimated from randomized alignments that are simulated along the same phylogenetic tree. The parameters of the extreme value distributed random scores are estimated and used to assign _P_-values to the observed segments in the native alignment.
FIGURE 2.
Examples of typical gap patterns and scoring paths in a pairwise alignment assumed to be coding. Nucleotides are shown as blocks, codons as three consecutive blocks of the same shading. (A) A gap of length three does not change the reading frame and in-frame-aligned codons are scored with the normalized substitution score σ. (B) A single gap destroys the reading frame but gets corrected downstream by another gap. The triplets that are out-of-phase because of this obvious alignment error are penalized by the two frameshift penalties Ω and ω. (C) A single gap that, in principle, destroys the reading frame is interpreted as a sequence error. Penalized by a high negative score Δ, this frameshift is ignored, and downstream codons are considered to be in-phase.
FIGURE 3.
RNAcode results on comparative test sets from various species. (A) Score distributions of annotated coding regions and randomly chosen noncoding regions in the Drosophila test set. (B) ROC curves for all six test sets. The full curve for all ranges of sensitivity/specificity from 0 to 1 is shown in the main diagrams. (Insets) The high specificity rate with false positive rates from 0 to 0.1. (C) Score distribution of noncoding alignments. The same distribution of the Drosophila test set as shown in A is shown in more detail. The fitted Gumbel distribution is shown as dotted line. (Upper right diagram) Comparison of the calculated _P_-values (via simulation and fitting of the Gumbel distribution) to the empirical _P_-values, i.e., the actual observed frequencies in the test set.
FIGURE 4.
Comparison of the RNAcode substitution score with other comparative metrics. The ROC curves show the classification performance of the dN/dS ratio, substitution rate variation, and the average substitution score σ used by RNAcode. Results are shown for alignments of length 30 from vertebrates, archaebacteria, yeasts, and drosophilid species grouped by the number of sequences in the alignment (N) and the mean pairwise sequence identity (MPI). The area under the ROC curve (AUC) as a measure for classification performance is shown for all methods and sets.
FIGURE 5.
Examples of novel short proteins in Escherichia coli. Sequence, genomic context, the high-scoring RNAcode segment, and fragment ion mass spectra are shown. Genome browser screenshots were made at
(Schneider et al. 2006). Arrows within annotated elements indicate their reading direction. The shading of mutational patterns was directly produced by the RNAcode program. The full species names for the abbreviations can be found in Supplemental Table 1. The mass spectra are shown for two selected proteolytic peptides, which were scored with 80% probability and used in combination with the detection of additional peptides to confirm the expression of the candidates (for details, see Supplemental Table 3). The proteins shown in A and B correspond to candidates 28 and 19, respectively, listed in Supplemental Tables 2 and 3.
FIGURE 6.
Examples of ambiguities between the coding and noncoding nature of three RNAs. (A) The RNA C0343 from E. coli is listed as a noncoding RNA in Rfam. However, it overlaps with an RNAcode-predicted coding segment. While there is no evidence for a RNA secondary structure according to the RNAz classification value, the highly significant RNAcode prediction and MS experiments suggest that C0343 is an mRNA and not an ncRNA. (B) RNAIII of Staphylococcus aureus (Rfam RF00503) contains a short ORF of a hemolysin gene. RNAcode predicts the open reading frame at the correct position, while RNAz clearly detects a structural signal. These results are consistent with the well-established dual nature of this molecule. (C) The Bacillus subtilis RNA SR1 is known to have function on the RNA level by targeting an mRNA. RNAcode detects a short ORF that was shown by Gimpel et al. (2010) to produce a small peptide and is thus another example of a dual-function RNA.
FIGURE 7.
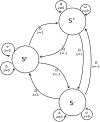
Finite state automaton representing the scoring of pairwise alignments. The three states correspond to the relative phases of the sequences. Insertions and deletions with z ≠ 0 lead to local changes in-phase that are penalized by Ω. Extension in each of the two out-of-frame states S+ and S− is penalized by ω. In/dels interpreted as sequencing errors or true frameshifts are penalized by Δ.
Similar articles
- RNAcode_Web - Convenient identification of evolutionary conserved protein coding regions.
Anders J, Stadler PF. Anders J, et al. J Integr Bioinform. 2023 Aug 25;20(3):20220046. doi: 10.1515/jib-2022-0046. eCollection 2023 Sep 1. J Integr Bioinform. 2023. PMID: 37615674 Free PMC article. - MASTR: multiple alignment and structure prediction of non-coding RNAs using simulated annealing.
Lindgreen S, Gardner PP, Krogh A. Lindgreen S, et al. Bioinformatics. 2007 Dec 15;23(24):3304-11. doi: 10.1093/bioinformatics/btm525. Epub 2007 Nov 15. Bioinformatics. 2007. PMID: 18006551 - RILogo: visualizing RNA-RNA interactions.
Menzel P, Seemann SE, Gorodkin J. Menzel P, et al. Bioinformatics. 2012 Oct 1;28(19):2523-6. doi: 10.1093/bioinformatics/bts461. Epub 2012 Jul 23. Bioinformatics. 2012. PMID: 22826541 - A practical guide to the art of RNA gene prediction.
Meyer IM. Meyer IM. Brief Bioinform. 2007 Nov;8(6):396-414. doi: 10.1093/bib/bbm011. Epub 2007 May 4. Brief Bioinform. 2007. PMID: 17483123 Review. - Methods for comprehensive experimental identification of RNA-protein interactions.
McHugh CA, Russell P, Guttman M. McHugh CA, et al. Genome Biol. 2014 Jan 27;15(1):203. doi: 10.1186/gb4152. Genome Biol. 2014. PMID: 24467948 Free PMC article. Review.
Cited by
- Transcripts with in silico predicted RNA structure are enriched everywhere in the mouse brain.
Seemann SE, Sunkin SM, Hawrylycz MJ, Ruzzo WL, Gorodkin J. Seemann SE, et al. BMC Genomics. 2012 May 31;13:214. doi: 10.1186/1471-2164-13-214. BMC Genomics. 2012. PMID: 22651826 Free PMC article. - The locality dilemma of Sankoff-like RNA alignments.
Müller T, Miladi M, Hutter F, Hofacker I, Will S, Backofen R. Müller T, et al. Bioinformatics. 2020 Jul 1;36(Suppl_1):i242-i250. doi: 10.1093/bioinformatics/btaa431. Bioinformatics. 2020. PMID: 32657398 Free PMC article. - An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions.
Haudry A, Platts AE, Vello E, Hoen DR, Leclercq M, Williamson RJ, Forczek E, Joly-Lopez Z, Steffen JG, Hazzouri KM, Dewar K, Stinchcombe JR, Schoen DJ, Wang X, Schmutz J, Town CD, Edger PP, Pires JC, Schumaker KS, Jarvis DE, Mandáková T, Lysak MA, van den Bergh E, Schranz ME, Harrison PM, Moses AM, Bureau TE, Wright SI, Blanchette M. Haudry A, et al. Nat Genet. 2013 Aug;45(8):891-8. doi: 10.1038/ng.2684. Epub 2013 Jun 30. Nat Genet. 2013. PMID: 23817568 - The Transcriptome of Exophiala dermatitidis during Ex-vivo Skin Model Infection.
Poyntner C, Blasi B, Arcalis E, Mirastschijski U, Sterflinger K, Tafer H. Poyntner C, et al. Front Cell Infect Microbiol. 2016 Oct 24;6:136. doi: 10.3389/fcimb.2016.00136. eCollection 2016. Front Cell Infect Microbiol. 2016. PMID: 27822460 Free PMC article. - Reekeekee- and roodoodooviruses, two different Microviridae clades constituted by the smallest DNA phages.
Olo Ndela E, Roux S, Henke C, Sczyrba A, Sime Ngando T, Varsani A, Enault F. Olo Ndela E, et al. Virus Evol. 2022 Dec 23;9(1):veac123. doi: 10.1093/ve/veac123. eCollection 2023. Virus Evol. 2022. PMID: 36694818 Free PMC article.
References
- Aebersold R, Mann M 2003. Mass spectrometry-based proteomics. Nature 422: 198–207 - PubMed
- Badger JH, Olsen GJ 1999. CRITICA: Coding region identification tool invoking comparative analysis. Mol Biol Evol 16: 512–524 - PubMed
- Bofkin L, Goldman N 2007. Variation in evolutionary processes at different codon positions. Mol Biol Evol 24: 513–521 - PubMed
- Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, et al. 2007. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev 21: 1353–1366 - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous