Analysis of the lung microbiome in the "healthy" smoker and in COPD - PubMed (original) (raw)
Clinical Trial
. 2011 Feb 22;6(2):e16384.
doi: 10.1371/journal.pone.0016384.
Deborah L Thompson, Meilan K Han, Christine M Freeman, Lisa McCloskey, Lindsay A Schmidt, Vincent B Young, Galen B Toews, Jeffrey L Curtis, Baskaran Sundaram, Fernando J Martinez, Gary B Huffnagle
Affiliations
- PMID: 21364979
- PMCID: PMC3043049
- DOI: 10.1371/journal.pone.0016384
Clinical Trial
Analysis of the lung microbiome in the "healthy" smoker and in COPD
John R Erb-Downward et al. PLoS One. 2011.
Abstract
Although culture-independent techniques have shown that the lungs are not sterile, little is known about the lung microbiome in chronic obstructive pulmonary disease (COPD). We used pyrosequencing of 16S amplicons to analyze the lung microbiome in two ways: first, using bronchoalveolar lavage (BAL) to sample the distal bronchi and air-spaces; and second, by examining multiple discrete tissue sites in the lungs of six subjects removed at the time of transplantation. We performed BAL on three never-smokers (NS) with normal spirometry, seven smokers with normal spirometry ("healthy smokers", HS), and four subjects with COPD (CS). Bacterial 16 s sequences were found in all subjects, without significant quantitative differences between groups. Both taxonomy-based and taxonomy-independent approaches disclosed heterogeneity in the bacterial communities between HS subjects that was similar to that seen in healthy NS and two mild COPD patients. The moderate and severe COPD patients had very limited community diversity, which was also noted in 28% of the healthy subjects. Both approaches revealed extensive membership overlap between the bacterial communities of the three study groups. No genera were common within a group but unique across groups. Our data suggests the existence of a core pulmonary bacterial microbiome that includes Pseudomonas, Streptococcus, Prevotella, Fusobacterium, Haemophilus, Veillonella, and Porphyromonas. Most strikingly, there were significant micro-anatomic differences in bacterial communities within the same lung of subjects with advanced COPD. These studies are further demonstration of the pulmonary microbiome and highlight global and micro-anatomic changes in these bacterial communities in severe COPD patients.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
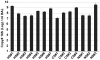
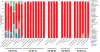
Figure 1. 16S qPCR of BAL Samples.
The number of copies of bacterial 16S per ml of BAL fluid was measured by qPCR (as described in Methods and Materials). The samples were divided into three groups: healthy smoker (HS), COPD subject (CS), and never-smoker (NS); the individual samples are displayed along the x-axis (Mean ± SEM). Samples where run in duplicate with two 10-fold dilutions.
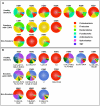
Figure 2. Taxonomic Classification of Bacterial Communities Present in the BAL.
The V1-V3 region of the bacterial 16S genes were sequenced using 454-pyrosequencing and taxonomically classified using RDP Classifier. A. Phylum level classification of the 16S amplicons present in a given subject. B. Genus level classification of these amplicons. The numbers at the bottom of each pie chart identify the organisms which can be found in Table 3.
Figure 3. Identification of Bacterial Community Membership Overlap in Subject BALs.
The relative abundance of each genera present in the BAL of each subject are plotted together with the numbers along the x-axis corresponding to the rank from Table 3. The study subject is displayed along the z-axis and the relative abundance (as a percent) is displayed along the y-axis. The genera Pseudomonas, Streptococcus, Prevotella, Fusobacterium, Veillonella and Prophyromonas are highlighted due to the dominance of these organisms within all of the subjects examined.
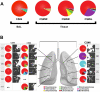
Figure 4. Bacterial Communities Present in Individual Lung Airways.
A. Bacterial community profiles for an entire explanted lung lobe from subjects with severe COPD. The total aggregate genus level reads of samples taken from the right lung of subject CS#5, the right lung of subject CS#6, and the left lung of subject CS#6 were analyzed on a per lung basis and compared to the BAL of subject CS#4 (reproduced from Figure 2). B. Multiple samples were taken from lung explants (right lung, subject CS#5; both lungs, subject CS#6) at the time of elective transplantation. Samples were harvested from the regions of lung indicated by the arrows on the gray lung schematic. Pie diagrams depict the genus level classification of 16S sequences, and the CT images demonstrate the absence of bronchiectasis in the airways adjacent to where samples were obtained. The key for the nine most abundant organisms is provided below the lung schematic. The full community breakdown for each of the airways can be found in Table S1.
Figure 5. Bacterial Distribution Throughout COPD Lung Explants.
Bacterial communities were characterized in the airways from 5 lobes of 4 lung explants. Multiple samples (4-8) were taken throughout the lung explants at the time of elective transplantation. The barchart depicts the genus level classification of 16S sequences identified.
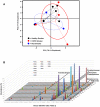
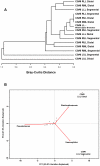
Figure 6. Cluster Analysis of the Bacterial Communities Sampled from Sites.
A. Furthest-neighbor joining tree built on the Bray-Curtis distance and B. Biplot of the principle components analysis of the normalized bacterial communities from multiple anatomic sites in the lung explants.
Similar articles
- The lung tissue microbiome in chronic obstructive pulmonary disease.
Sze MA, Dimitriu PA, Hayashi S, Elliott WM, McDonough JE, Gosselink JV, Cooper J, Sin DD, Mohn WW, Hogg JC. Sze MA, et al. Am J Respir Crit Care Med. 2012 May 15;185(10):1073-80. doi: 10.1164/rccm.201111-2075OC. Epub 2012 Mar 15. Am J Respir Crit Care Med. 2012. PMID: 22427533 Free PMC article. - Lung microbiota associations with clinical features of COPD in the SPIROMICS cohort.
Opron K, Begley LA, Erb-Downward JR, Freeman C, Madapoosi S, Alexis NE, Barjaktarevic I, Graham Barr R, Bleecker ER, Bowler RP, Christenson SA, Comellas AP, Cooper CB, Couper DJ, Doerschuk CM, Dransfield MT, Han MK, Hansel NN, Hastie AT, Hoffman EA, Kaner RJ, Krishnan J, O'Neal WK, Ortega VE, Paine R 3rd, Peters SP, Michael Wells J, Woodruff PG, Martinez FJ, Curtis JL, Huffnagle GB, Huang YJ. Opron K, et al. NPJ Biofilms Microbiomes. 2021 Feb 5;7(1):14. doi: 10.1038/s41522-021-00185-9. NPJ Biofilms Microbiomes. 2021. PMID: 33547327 Free PMC article. - Analysis of the airway microbiota of healthy individuals and patients with chronic obstructive pulmonary disease by T-RFLP and clone sequencing.
Zakharkina T, Heinzel E, Koczulla RA, Greulich T, Rentz K, Pauling JK, Baumbach J, Herrmann M, Grünewald C, Dienemann H, von Müller L, Bals R. Zakharkina T, et al. PLoS One. 2013 Jul 9;8(7):e68302. doi: 10.1371/journal.pone.0068302. Print 2013. PLoS One. 2013. PMID: 23874580 Free PMC article. - COPD and the microbiome.
Mammen MJ, Sethi S. Mammen MJ, et al. Respirology. 2016 May;21(4):590-9. doi: 10.1111/resp.12732. Epub 2016 Jan 27. Respirology. 2016. PMID: 26852737 Review. - Role of the Lung Microbiome in the Pathogenesis of Chronic Obstructive Pulmonary Disease.
Wang L, Hao K, Yang T, Wang C. Wang L, et al. Chin Med J (Engl). 2017 Sep 5;130(17):2107-2111. doi: 10.4103/0366-6999.211452. Chin Med J (Engl). 2017. PMID: 28741603 Free PMC article. Review.
Cited by
- Exploring the potential role of microbiota and metabolites in acute exacerbation of chronic obstructive pulmonary disease.
Shi Y, Yang J, Tian T, Li S, Xie Y. Shi Y, et al. Front Microbiol. 2024 Oct 16;15:1487393. doi: 10.3389/fmicb.2024.1487393. eCollection 2024. Front Microbiol. 2024. PMID: 39483760 Free PMC article. - The role of the lung microbiome in health and disease. A National Heart, Lung, and Blood Institute workshop report.
Huang YJ, Charlson ES, Collman RG, Colombini-Hatch S, Martinez FD, Senior RM. Huang YJ, et al. Am J Respir Crit Care Med. 2013 Jun 15;187(12):1382-7. doi: 10.1164/rccm.201303-0488WS. Am J Respir Crit Care Med. 2013. PMID: 23614695 Free PMC article. - Data Mining of Lung Microbiota in Cystic Fibrosis Patients.
Li J, Hao C, Ren L, Xiao Y, Wang J, Qin X. Li J, et al. PLoS One. 2016 Oct 14;11(10):e0164510. doi: 10.1371/journal.pone.0164510. eCollection 2016. PLoS One. 2016. PMID: 27741283 Free PMC article. - Smoking-induced microbial dysbiosis in health and disease.
Shapiro H, Goldenberg K, Ratiner K, Elinav E. Shapiro H, et al. Clin Sci (Lond). 2022 Sep 30;136(18):1371-1387. doi: 10.1042/CS20220175. Clin Sci (Lond). 2022. PMID: 36156126 Free PMC article. Review. - The dynamic lung microbiome in health and disease.
Natalini JG, Singh S, Segal LN. Natalini JG, et al. Nat Rev Microbiol. 2023 Apr;21(4):222-235. doi: 10.1038/s41579-022-00821-x. Epub 2022 Nov 16. Nat Rev Microbiol. 2023. PMID: 36385637 Free PMC article. Review.
References
- Murray CJ, Lopez AD. Evidence-based health policy–lessons from the Global Burden of Disease Study. Science. 1996;274:740–743. - PubMed
- Lopez AD, Shibuya K, Rao C, Mathers CD, Hansell AL, et al. Chronic obstructive pulmonary disease: current burden and future projections. Eur Respir J. 2006;27:397–412. - PubMed
- Barnes P, Drazen J, Rennard S, Thomsom N. 2008. Asthma and COPD: Basic Mechanisms and Clinical Management.
- Salvi SS, Barnes PJ. Chronic obstructive pulmonary disease in non-smokers. Lancet. 2009;374:733–743. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 HL007749/HL/NHLBI NIH HHS/United States
- P30 CA46952/CA/NCI NIH HHS/United States
- N01 HR046162/HR/NHLBI NIH HHS/United States
- T32 HL07749/HL/NHLBI NIH HHS/United States
- UL1 RR024986/RR/NCRR NIH HHS/United States
- R01 DK070875/DK/NIDDK NIH HHS/United States
- R01 HL082480/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous