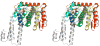Structure of the C-terminal domain of Saccharomyces cerevisiae Nup133, a component of the nuclear pore complex - PubMed (original) (raw)
doi: 10.1002/prot.22973. Epub 2011 Mar 1.
Tarun Gheyi, Stacy A Miller, Kevin T Bain, Mark Dickey, Jeffrey B Bonanno, Seung Joong Kim, Jeremy Phillips, Ursula Pieper, Javier Fernandez-Martinez, Josef D Franke, Anne Martel, Hiro Tsuruta, Shane Atwell, Devon A Thompson, J Spencer Emtage, Stephen R Wasserman, Michael P Rout, Andrej Sali, J Michael Sauder, Stephen K Burley
Affiliations
- PMID: 21365675
- PMCID: PMC3350809
- DOI: 10.1002/prot.22973
Structure of the C-terminal domain of Saccharomyces cerevisiae Nup133, a component of the nuclear pore complex
Parthasarathy Sampathkumar et al. Proteins. 2011 May.
Abstract
Nuclear pore complexes (NPCs), responsible for the nucleo-cytoplasmic exchange of proteins and nucleic acids, are dynamic macromolecular assemblies forming an eight-fold symmetric co-axial ring structure. Yeast (Saccharomyces cerevisiae) NPCs are made up of at least 456 polypeptide chains of ~30 distinct sequences. Many of these components (nucleoporins, Nups) share similar structural motifs and form stable subcomplexes. We have determined a high-resolution crystal structure of the C-terminal domain of yeast Nup133 (ScNup133), a component of the heptameric Nup84 subcomplex. Expression tests yielded ScNup133(944-1157) that produced crystals diffracting to 1.9Å resolution.
ScNup133(944-1157) adopts essentially an all α-helical fold, with a short two stranded β-sheet at the C-terminus. The 11 α-helices of ScNup133(944-1157) form a compact fold. In contrast, the previously determined structure of human Nup133(934-1156) bound to a fragment of human Nup107 has its constituent α-helices are arranged in two globular blocks. These differences may reflect structural divergence among homologous nucleoporins.
Figures
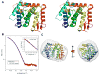
Figure 1
A: Stereo view of a ribbon representation of ScNup133(944-1157). Secondary structure elements were assigned by DSSP. The fold is colored blue to red from the N- to C-termini. B: Comparison of the merged experimental SAXS profile (red) of ScNup133(946-1157) with SAXS profiles computed by IMP-FoXS , (blue) for the complete model of ScNup133(944-1157). Inset shows the SAXS profiles in the Guinier plot. Dmax of radial distribution function, P(r) is 56.25Å). C: The shape of ScNup133(944-1157) derived from the experimental SAXS profile, shown in two orthogonal orientations.
Figure 2
A: Structural superposition of the C-terminal domains of Sc and Hs Nup133. ScNup133(944-1157) is shown as in Fig. 1 (A) and HuNup133(934-1156) is shown in grey.
Similar articles
- Integrative structure-function mapping of the nucleoporin Nup133 suggests a conserved mechanism for membrane anchoring of the nuclear pore complex.
Kim SJ, Fernandez-Martinez J, Sampathkumar P, Martel A, Matsui T, Tsuruta H, Weiss TM, Shi Y, Markina-Inarrairaegui A, Bonanno JB, Sauder JM, Burley SK, Chait BT, Almo SC, Rout MP, Sali A. Kim SJ, et al. Mol Cell Proteomics. 2014 Nov;13(11):2911-26. doi: 10.1074/mcp.M114.040915. Epub 2014 Aug 19. Mol Cell Proteomics. 2014. PMID: 25139911 Free PMC article. - Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145.
Sampathkumar P, Ozyurt SA, Do J, Bain KT, Dickey M, Rodgers LA, Gheyi T, Sali A, Kim SJ, Phillips J, Pieper U, Fernandez-Martinez J, Franke JD, Martel A, Tsuruta H, Atwell S, Thompson DA, Emtage JS, Wasserman SR, Rout MP, Sauder JM, Burley SK. Sampathkumar P, et al. Proteins. 2010 Jun;78(8):1992-8. doi: 10.1002/prot.22707. Proteins. 2010. PMID: 20310066 Free PMC article. No abstract available. - Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex.
Sampathkumar P, Kim SJ, Upla P, Rice WJ, Phillips J, Timney BL, Pieper U, Bonanno JB, Fernandez-Martinez J, Hakhverdyan Z, Ketaren NE, Matsui T, Weiss TM, Stokes DL, Sauder JM, Burley SK, Sali A, Rout MP, Almo SC. Sampathkumar P, et al. Structure. 2013 Apr 2;21(4):560-71. doi: 10.1016/j.str.2013.02.005. Epub 2013 Mar 14. Structure. 2013. PMID: 23499021 Free PMC article. - Toward the atomic structure of the nuclear pore complex: when top down meets bottom up.
Hoelz A, Glavy JS, Beck M. Hoelz A, et al. Nat Struct Mol Biol. 2016 Jul;23(7):624-30. doi: 10.1038/nsmb.3244. Epub 2016 Jun 6. Nat Struct Mol Biol. 2016. PMID: 27273515 Free PMC article. Review. - Nuclear pore complexes: round the bend?
Antonin W, Mattaj IW. Antonin W, et al. Nat Cell Biol. 2005 Jan;7(1):10-2. doi: 10.1038/ncb0105-10. Nat Cell Biol. 2005. PMID: 15632943 Review. No abstract available.
Cited by
- Evolutionary divergence of the nuclear pore complex from fungi to metazoans.
Chopra K, Bawaria S, Chauhan R. Chopra K, et al. Protein Sci. 2019 Mar;28(3):571-586. doi: 10.1002/pro.3558. Epub 2018 Dec 24. Protein Sci. 2019. PMID: 30488506 Free PMC article. - Structure-function mapping of a heptameric module in the nuclear pore complex.
Fernandez-Martinez J, Phillips J, Sekedat MD, Diaz-Avalos R, Velazquez-Muriel J, Franke JD, Williams R, Stokes DL, Chait BT, Sali A, Rout MP. Fernandez-Martinez J, et al. J Cell Biol. 2012 Feb 20;196(4):419-34. doi: 10.1083/jcb.201109008. Epub 2012 Feb 13. J Cell Biol. 2012. PMID: 22331846 Free PMC article. - Integrative structure-function mapping of the nucleoporin Nup133 suggests a conserved mechanism for membrane anchoring of the nuclear pore complex.
Kim SJ, Fernandez-Martinez J, Sampathkumar P, Martel A, Matsui T, Tsuruta H, Weiss TM, Shi Y, Markina-Inarrairaegui A, Bonanno JB, Sauder JM, Burley SK, Chait BT, Almo SC, Rout MP, Sali A. Kim SJ, et al. Mol Cell Proteomics. 2014 Nov;13(11):2911-26. doi: 10.1074/mcp.M114.040915. Epub 2014 Aug 19. Mol Cell Proteomics. 2014. PMID: 25139911 Free PMC article. - Yeast Nup84-Nup133 complex structure details flexibility and reveals conservation of the membrane anchoring ALPS motif.
Nordeen SA, Turman DL, Schwartz TU. Nordeen SA, et al. Nat Commun. 2020 Nov 27;11(1):6060. doi: 10.1038/s41467-020-19885-5. Nat Commun. 2020. PMID: 33247142 Free PMC article. - The Structure of the Nuclear Pore Complex (An Update).
Lin DH, Hoelz A. Lin DH, et al. Annu Rev Biochem. 2019 Jun 20;88:725-783. doi: 10.1146/annurev-biochem-062917-011901. Epub 2019 Mar 18. Annu Rev Biochem. 2019. PMID: 30883195 Free PMC article. Review.
References
- Elizabeth JT, Wente SR. Dynamic nuclear pore complexes: Life on the edge. Cell. 2006;125:1041–1053. - PubMed
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang WH, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Sali A, Rout MP. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. - PubMed
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang WZ, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Rout MP, Sali A. Determining the architectures of macromolecular assemblies. Nature. 2007;450:683–694. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54 GM074945-05/GM/NIGMS NIH HHS/United States
- U54 RR022220-07/RR/NCRR NIH HHS/United States
- R01 GM083960/GM/NIGMS NIH HHS/United States
- U54 GM074945/GM/NIGMS NIH HHS/United States
- U54 RR022220/RR/NCRR NIH HHS/United States
- R01 GM083960-04/GM/NIGMS NIH HHS/United States
- R01 GM062427-11/GM/NIGMS NIH HHS/United States
- R01 GM062427/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous