Chaperone role for proteins p618 and p892 in the extracellular tail development of Acidianus two-tailed virus - PubMed (original) (raw)
Chaperone role for proteins p618 and p892 in the extracellular tail development of Acidianus two-tailed virus
Urte Scheele et al. J Virol. 2011 May.
Abstract
The crenarchaeal Acidianus two-tailed virus (ATV) undergoes a remarkable morphological development, extracellularly and independently of host cells, by growing long tails at each end of a spindle-shaped virus particle. Initial work suggested that an intermediate filament-like protein, p800, is involved in this process. We propose that an additional chaperone system is required, consisting of a MoxR-type AAA ATPase (p618) and a von Willebrand domain A (VWA)-containing cochaperone, p892. Both proteins are absent from the other known bicaudavirus, STSV1, which develops a single tail intracellularly. p618 exhibits ATPase activity and forms a hexameric ring complex that closely resembles the oligomeric complex of the MoxR-like protein RavA (YieN). ATV proteins p387, p653, p800, and p892 interact with p618, and with the exception of p800, all bind to DNA. A model is proposed to rationalize the interactions observed between the different protein and DNA components and to explain their possible structural and functional roles in extracellular tail development.
Figures
Fig. 1.
Properties of ATV virion proteins. (A) Domain structures of p618, p892, p387, and p653. Structural domains and sequence motifs are color coded as follows: blue, AAA domain; green, disordered regions; light green, VWA domain; yellow, low-complexity regions; purple, coiled-coil elements; and light blue, shared domain of unknown function in p387 and p653. (B) Sequence alignment of p618 and RavA showing degrees of similarity. Conserved residues within the RavA subfamily (25) are shown in purple.
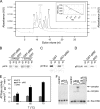
Fig. 2.
Properties of p618. (A) Measurement of ATPase activity. p618 (0.01 mg/ml) was incubated at 60°C with increasing amounts of ATP. Release of inorganic phosphate was determined by malachite green assay. The inset shows a Hill plot, with a Hill coefficient of 1. (B) Temperature dependence of ATPase activity was analyzed in the presence of 1 mM ATP at the indicated temperatures. (C to E) Oligomerization of p618 was analyzed by size exclusion chromatography of 60 μM p618 (C), 50 μM p618E128Q (D), and 12 μM p618K62A (E) in the presence or absence of 1 mM ATP monitored at 280 nm. (F) Coprecipitation experiments were performed with GST-p618 immobilized on GSH-Sepharose beads (GST-p618) and 0.2 mg/ml of p618ΔN in the presence of 1 mM ATP. GST conjugated to GSH beads (GST) and 0.25 mg/ml BSA served as controls. Samples (7.5 μl) of the supernatant (S) and pellet (P) fractions were analyzed by Western blotting using a penta-His antibody as primary antibody.
Fig. 3.
Hexameric ring structure of p618E128Q. Oligomeric complexes of the Walker B mutant of p618 were isolated by size exclusion chromatography at 4°C in the presence of 0.2 mM ATP and analyzed by electron microscopy after negative staining with uranyl acetate. The inset shows a detailed view of a hexameric ring.
Fig. 4.
Oligomerization and protein-protein interactions of p892. (A) Oligomerization of p892. The oligomeric state of p892 was analyzed by size exclusion chromatography and monitored at 280 nm. Thyroglobulin (T), ferritin (F), catalase (C), aldolase (A), and BSA (B) were used as standard proteins. Molecular masses of 640 kDa and 460 kDa were estimated from the calibration curve (inset) for the first and second elution peaks, consistent with hexamer and tetramer formation, respectively. (B to D) p892 interactions with p618. Deletion mutants of p618 lacking either the C-terminal (p618ΔC) or N-terminal (p618ΔN) domain were used to map the binding site on p618. Coprecipitation experiments were performed with GST-p892 immobilized on GSH-Sepharose beads (GST-p892) and 0.2 mg/ml of p618 (B), p618ΔC (C), and p618ΔN (D) in the presence of 1 mM ATP. GST conjugated to GSH beads (GST) and 0.25 mg/ml BSA served as controls. Supernatant (S) and pellet (P) fractions were analyzed by Western blotting. (E) The effect of p892 on the ATPase activity of p618 was measured at 650 nm. The ATPase assay was performed at the indicated temperatures in the presence of 1 mM ATP and a 5-fold molar excess of p892. p892 stimulated ATPase activity of p618 by 3.6-fold at 75°C. (F) Binding of p618 (0 to 20 μM), p892 (6 and 12 μM), and the p618-p892 complex to DNA was investigated at 50°C for 25 min with 1.5 nmol of labeled DNA (147 bp). p618 was added to p892 in an equimolar amount and a 2-fold molar excess in the last two lanes. The negative control contained substrate DNA only. Protein-bound and free DNAs were separated by electrophoresis on an 11% nondenaturing acrylamide gel and were visualized by autoradiography.
Fig. 5.
p387 protein-protein interactions. (A) Coprecipitation experiments were carried out with p387 and GST-p618. (B) The binding site of p387 on p618 was mapped using a p618 fragment lacking the N-terminal domain (p618ΔN). Assays were performed with 0.2 mg/ml of target proteins, and GST conjugated to GSH beads and BSA served as controls. (C) A competition pulldown experiment with p618 and p892 and with increasing concentrations of p387 (0.1 to 0.3 mg/ml) was performed as described above. Supernatant (S) and pellet (P) fractions were analyzed either by SDS-PAGE with Coomassie staining or by Western blotting (A and B). A molecular size standard (MW) was used to estimate the apparent molecular protein masses. (D) The effect of a 5-fold molar excess of p387 on the ATPase activity of p618 was estimated at 650 nm at the indicated temperatures in the presence of 1 mM ATP. (E and F) Electrophoretic mobility shift assays. (E) DNA binding of p387 was investigated at 50°C for 25 min with 1.5 nM DNA (147 bp) and increasing concentrations of protein (0 to 5 μM p387). (F) Binding of p618 to the DNA-p387 complex (molar ratio of p618 to p387, 0.5 to 2) resulted in the formation of a multimolecular complex. The negative controls contained substrate DNA only. Protein-bound and free DNAs were separated by electrophoresis on an 11% nondenaturing acrylamide gel and were visualized by autoradiography.
Fig. 6.
p653 protein and DNA interactions. (A) Coprecipitation experiments were performed with GST-p618 beads and 0.2 mg/ml of p653, using GST beads and BSA as controls. Supernatant (S) and pellet (P) fractions were analyzed by Western blotting. (B) DNA binding of p653 was investigated at 50°C for 25 min with 1.5 nM DNA (147 bp) and 0 to 2.5 μM p653. DNA without protein served as a negative control. Protein-bound and free DNAs were separated by electrophoresis on an 11% nondenaturing acrylamide gel and were visualized by autoradiography.
Fig. 7.
Assembly and protein-protein interactions of p800. (A) Temperature dependence of the assembly of p800 into filaments. p800 was purified by size exclusion chromatography, kept overnight at 4°C, and then incubated for 5 min at either 22°C or 50°C. The filaments were analyzed by electron microscopy after negative staining with uranyl acetate. (B and C) Interaction of p800 with p618 and p387. Coprecipitation experiments with GST-p618 (B) and GST-p387 (C) were performed with 0.3 mg/ml p800. GST beads and BSA were used as controls. Supernatants (S) and beads as the pellet fraction (P) were analyzed by Western blotting.
Similar articles
- Structural and genomic properties of the hyperthermophilic archaeal virus ATV with an extracellular stage of the reproductive cycle.
Prangishvili D, Vestergaard G, Häring M, Aramayo R, Basta T, Rachel R, Garrett RA. Prangishvili D, et al. J Mol Biol. 2006 Jun 23;359(5):1203-16. doi: 10.1016/j.jmb.2006.04.027. Epub 2006 Apr 27. J Mol Biol. 2006. PMID: 16677670 - AAA ATPase p529 of Acidianus two-tailed virus ATV and host receptor recognition.
Erdmann S, Scheele U, Garrett RA. Erdmann S, et al. Virology. 2011 Dec 5;421(1):61-6. doi: 10.1016/j.virol.2011.08.029. Epub 2011 Oct 7. Virology. 2011. PMID: 21982819 - Novel structural and functional insights into the MoxR family of AAA+ ATPases.
Wong KS, Houry WA. Wong KS, et al. J Struct Biol. 2012 Aug;179(2):211-21. doi: 10.1016/j.jsb.2012.03.010. Epub 2012 Apr 3. J Struct Biol. 2012. PMID: 22491058 Review. - Crystal structure of ATV(ORF273), a new fold for a thermo- and acido-stable protein from the Acidianus two-tailed virus.
Felisberto-Rodrigues C, Blangy S, Goulet A, Vestergaard G, Cambillau C, Garrett RA, Ortiz-Lombardía M. Felisberto-Rodrigues C, et al. PLoS One. 2012;7(10):e45847. doi: 10.1371/journal.pone.0045847. Epub 2012 Oct 8. PLoS One. 2012. PMID: 23056221 Free PMC article. - Molecular chaperones HscA/Ssq1 and HscB/Jac1 and their roles in iron-sulfur protein maturation.
Vickery LE, Cupp-Vickery JR. Vickery LE, et al. Crit Rev Biochem Mol Biol. 2007 Mar-Apr;42(2):95-111. doi: 10.1080/10409230701322298. Crit Rev Biochem Mol Biol. 2007. PMID: 17453917 Review.
Cited by
- Highly regulated, diversifying NTP-dependent biological conflict systems with implications for the emergence of multicellularity.
Kaur G, Burroughs AM, Iyer LM, Aravind L. Kaur G, et al. Elife. 2020 Feb 26;9:e52696. doi: 10.7554/eLife.52696. Elife. 2020. PMID: 32101166 Free PMC article. - Identification and characterization of multiple rubisco activases in chemoautotrophic bacteria.
Tsai YC, Lapina MC, Bhushan S, Mueller-Cajar O. Tsai YC, et al. Nat Commun. 2015 Nov 16;6:8883. doi: 10.1038/ncomms9883. Nat Commun. 2015. PMID: 26567524 Free PMC article. - Jumbo Phages: A Comparative Genomic Overview of Core Functions and Adaptions for Biological Conflicts.
M Iyer L, Anantharaman V, Krishnan A, Burroughs AM, Aravind L. M Iyer L, et al. Viruses. 2021 Jan 5;13(1):63. doi: 10.3390/v13010063. Viruses. 2021. PMID: 33466489 Free PMC article. - A novel single-tailed fusiform Sulfolobus virus STSV2 infecting model Sulfolobus species.
Erdmann S, Chen B, Huang X, Deng L, Liu C, Shah SA, Le Moine Bauer S, Sobrino CL, Wang H, Wei Y, She Q, Garrett RA, Huang L, Lin L. Erdmann S, et al. Extremophiles. 2014 Jan;18(1):51-60. doi: 10.1007/s00792-013-0591-z. Epub 2013 Oct 27. Extremophiles. 2014. PMID: 24163004 - Viruses of haloarchaea.
Luk AW, Williams TJ, Erdmann S, Papke RT, Cavicchioli R. Luk AW, et al. Life (Basel). 2014 Nov 13;4(4):681-715. doi: 10.3390/life4040681. Life (Basel). 2014. PMID: 25402735 Free PMC article. Review.
References
- Bamford D. H., Caldentey J., Bamford J. K. 1995. Bacteriophage PRD1: a broad host range DSDNA tectivirus with an internal membrane. Adv. Virus Res. 45:281–319 - PubMed
- Bosl B., Grimminger V., Walter S. 2005. Substrate binding to the molecular chaperone Hsp104 and its regulation by nucleotides. J. Biol. Chem. 280:38170–38176 - PubMed
- DeLaBarre B., Brunger A. T. 2005. Nucleotide dependent motion and mechanism of action of p97/VCP. J. Mol. Biol. 347:437–452 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials