Isolation of MECP2-null Rett Syndrome patient hiPS cells and isogenic controls through X-chromosome inactivation - PubMed (original) (raw)
. 2011 Jun 1;20(11):2103-15.
doi: 10.1093/hmg/ddr093. Epub 2011 Mar 3.
Affiliations
- PMID: 21372149
- PMCID: PMC3090191
- DOI: 10.1093/hmg/ddr093
Isolation of MECP2-null Rett Syndrome patient hiPS cells and isogenic controls through X-chromosome inactivation
Aaron Y L Cheung et al. Hum Mol Genet. 2011.
Abstract
Rett syndrome (RTT) is a neurodevelopmental autism spectrum disorder that affects girls due primarily to mutations in the gene encoding methyl-CpG binding protein 2 (MECP2). The majority of RTT patients carry missense and nonsense mutations leading to a hypomorphic MECP2, while null mutations leading to the complete absence of a functional protein are rare. MECP2 is an X-linked gene subject to random X-chromosome inactivation resulting in mosaic expression of mutant MECP2. The lack of human brain tissue motivates the need for alternative human cellular models to study RTT. Here we report the characterization of a MECP2 mutation in a classic female RTT patient involving rearrangements that remove exons 3 and 4 creating a functionally null mutation. To generate human neuron models of RTT, we isolated human induced pluripotent stem (hiPS) cells from RTT patient fibroblasts. RTT-hiPS cells retained the MECP2 mutation, are pluripotent and fully reprogrammed, and retained an inactive X-chromosome in a nonrandom pattern. Taking advantage of the latter characteristic, we obtained a pair of isogenic wild-type and mutant MECP2 expressing RTT-hiPS cell lines that retained this MECP2 expression pattern upon differentiation into neurons. Phenotypic analysis of mutant RTT-hiPS cell-derived neurons demonstrated a reduction in soma size compared with the isogenic control RTT-hiPS cell-derived neurons from the same RTT patient. Analysis of isogenic control and mutant hiPS cell-derived neurons represents a promising source for understanding the pathogenesis of RTT and the role of MECP2 in human neurons.
Figures
Figure 1.
Mapping of the Δ3–4 MECP2 mutation. (A) Schematic of the MECP2 locus. Primers used for analysis of copy number variations and their approximate amplifying regions are indicated, where applicable, with orange bars. Primers in italics belong to intron 2. Primers in dark blue were used for amplification of WT (KR6-Fwd and KR16-Rev) and mutant Δ3–4 (AC7-Fwd and KR16-Rev) MECP2 alleles in (C) and their approximate location are indicated with dark blue arrows. The two alleles of MECP2 are indicated with red bars. There are two deletions that comprise the Δ3–4 MECP2 mutation, g.61340_67032delinsAGTTGTGCCAC and g.67072_67200del. The pair of deletions is indicated by the absence of one of the red bars indicative of the deletion of one of the two alleles creating a heterozygous deletion. The insertion associated with the larger deletion is indicated with a light blue bar. The nomenclature of the mutations relates to the genomic DNA sequence with position 1 defined as the first nucleotide of NG_007107.1 (NCBI: reference sequence). The approximate location of the p.T158M and p.R306C mutations is indicated. MBD, methyl-CpG binding domain; TRD, transcriptional repression domain; UTR, untranslated region. (B) qPCR analysis of copy number variations along the MECP2 locus. Data are expressed as mean ± SEM. (C) PCR using WT and mutant-specific primers to amplify the WT and Δ3–4 MECP2 allele, respectively.
Figure 2.
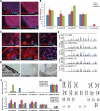
Δ3–4-hiPS cells are pluripotent and fully reprogrammed. (A) Δ3–4-hiPS #37 express pluripotency markers by immunocytochemistry. Scale bars, 100 µm. (B) Δ3–4-hiPS cells express bona fide hiPS cell markers by qRT–PCR. Data are expressed as mean ± SEM. (C) Δ3–4-hiPS #37 differentiate into the three germ layers, ectoderm (TUJ1, NESTIN), mesoderm [MYOSIN, SMA (SMOOTH MUSCLE ACTIN)] and endoderm (GATA4, SOX17), in vitro via embryoid body formation. Scale bars, 50 µm. (D) Δ3–4-hiPS #37 differentiates into the three germ layers, ectoderm (pigmented epithelium), mesoderm (cartilage) and endoderm (gut epithelium), in vivo via teratoma formation by injection into immunodeficient mice. Scale bars, 50 µm. (E) Δ3–4-hiPS cells carry identical short tandem repeat profile as their parental fibroblast of origin and are distinct from CA1 hES cells in the laboratory. (F) qRT–PCR shows that Δ3–4-hiPS cells have largely silenced the reprogramming vectors in comparison to IMR90-4F+RFP and have reactivated the endogenous loci of the reprogramming factors similar to H9 hES cells. ‘pMXs' and ‘Endo' refers to primers specifically recognizing the exogenous reprogramming factors and endogenous loci, respectively. Data are expressed as mean ± SEM. (G) Δ3–4-hiPS #37 carries a normal female karyotype by G-banding analysis.
Figure 3.
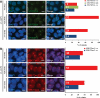
Female Δ3–4-hiPS cells retain an inactive X-chromosome. Δ3–4-hiPS cells were assessed by RNA-FISH and immunocytochemistry for XIST RNA (A) and H3K27me3 (B), respectively. Graph depicts the percentage of colonies with a single XIST RNA (A) and H3K27me3 (B) signal. Positive (+ve) colonies indicate that >90% of the cells observed within the colony were positive for the signal. Examples of positive colonies are shown on the left panels. Negative (−ve) colonies indicate that no cells within the colony were positive for the signal. Mix colonies indicate colonies where a single XIST RNA signal was detectable, but were below the threshold for a positive colony. The detection of these XCI marks suggests that Δ3–4-hiPS cells retain an inactive X-chromosome, at least in a large fraction of cells. The number of colonies analysed is indicated within each bar. Passage number of analysis is indicated in brackets. P, passage. Scale bars, 10 µm.
Figure 4.
XCI is nonrandom in female RTT-hiPS cells. RTT-fibroblasts and -hiPS cells were assessed for XCI patterns via the AR assay. Δ3–4- (A), T158M- (B), and R306C- (C) fibroblasts exhibited a random XCI pattern as shown by the presence of two different-sized amplicons, 174 and 180 bp, 168 and 177 bp, and 171 and 174 bp, respectively, after digestion with methylation-sensitive enzymes. Δ3–4- (A), T158M- (B), and R306C- (C) hiPS cells exhibited an extreme XCI skewing pattern as shown by the preferential detection of a single-sized peak after digestion with methylation-sensitive enzymes. The corrected XCI ratio (see Materials and Methods) is indicated on the top left of the digested graph.
Figure 5.
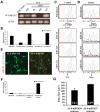
MECP2 expression follows the pattern of XCI in RTT-hiPS cells and their neuronal derivatives. RT–PCR (A) and qRT–PCR (B) of Δ3–4-hiPS cells detected WT MECP2 transcripts in Δ3–4-hiPS #6 and #37 but not in Δ3–4-hiPS #20, indicating Δ3–4-hiPS #20 expresses the Δ3–4 mutant MECP2 transcript. Diff. fNSCs, differentiated fetal neural stem cells; Fib, fibroblasts. Data are expressed as mean ± SEM (B). Sequencing of T158M- (C) and R306C- (D) fibroblasts' cDNA revealed that the fibroblasts express a mixture of WT and mutant MECP2 transcripts as indicated by the ‘C' and ‘T' nucleotide peaks, respectively. Sequencing of T158M- (C) and R306C- (D) hiPS cells revealed that they only express the WT and mutant MECP2 transcripts, respectively. Mut, Mutant. Δ3–4-hiPS #20 and #37 were differentiated into neurons for 9–10 weeks. WT MECP2 expression was assessed by immunocytochemistry (E) and qRT–PCR (F) and was detected in Δ3–4-hiPS #37-derived neurons but not in Δ3–4-hiPS #20-derived neurons. Scale bars, 39 µm (E). Diff. fNSCs, differentiated fetal neural stem cells; Fib, fibroblasts. Data are expressed as mean ± SEM (F). (G) Δ3–4-hiPS #20 and #37 were differentiated into neurons for 9 weeks. Δ3–4-hiPS #20-derived neurons exhibited significant reduction in soma size compared with Δ3–4-hiPS #37-derived neurons (*, P < 0.0001, Student's _t_-test). Total number of neurons (n) analysed per hiPS cell line from two independent biological replicates is indicated at the bottom of each bar. Data are expressed as mean ± SEM.
Similar articles
- Isogenic pairs of wild type and mutant induced pluripotent stem cell (iPSC) lines from Rett syndrome patients as in vitro disease model.
Ananiev G, Williams EC, Li H, Chang Q. Ananiev G, et al. PLoS One. 2011;6(9):e25255. doi: 10.1371/journal.pone.0025255. Epub 2011 Sep 26. PLoS One. 2011. PMID: 21966470 Free PMC article. - Neuronal maturation defect in induced pluripotent stem cells from patients with Rett syndrome.
Kim KY, Hysolli E, Park IH. Kim KY, et al. Proc Natl Acad Sci U S A. 2011 Aug 23;108(34):14169-74. doi: 10.1073/pnas.1018979108. Epub 2011 Aug 1. Proc Natl Acad Sci U S A. 2011. PMID: 21807996 Free PMC article. - Differentiation of multipotent neural stem cells derived from Rett syndrome patients is biased toward the astrocytic lineage.
Andoh-Noda T, Akamatsu W, Miyake K, Matsumoto T, Yamaguchi R, Sanosaka T, Okada Y, Kobayashi T, Ohyama M, Nakashima K, Kurosawa H, Kubota T, Okano H. Andoh-Noda T, et al. Mol Brain. 2015 May 27;8:31. doi: 10.1186/s13041-015-0121-2. Mol Brain. 2015. PMID: 26012557 Free PMC article. - Two new Rett syndrome families and review of the literature: expanding the knowledge of MECP2 frameshift mutations.
Ravn K, Roende G, Duno M, Fuglsang K, Eiklid KL, Tümer Z, Nielsen JB, Skjeldal OH. Ravn K, et al. Orphanet J Rare Dis. 2011 Aug 30;6:58. doi: 10.1186/1750-1172-6-58. Orphanet J Rare Dis. 2011. PMID: 21878110 Free PMC article. Review. - Rett syndrome: the complex nature of a monogenic disease.
Renieri A, Meloni I, Longo I, Ariani F, Mari F, Pescucci C, Cambi F. Renieri A, et al. J Mol Med (Berl). 2003 Jun;81(6):346-54. doi: 10.1007/s00109-003-0444-9. Epub 2003 May 16. J Mol Med (Berl). 2003. PMID: 12750821 Review.
Cited by
- Probing disorders of the nervous system using reprogramming approaches.
Ichida JK, Kiskinis E. Ichida JK, et al. EMBO J. 2015 Jun 3;34(11):1456-77. doi: 10.15252/embj.201591267. Epub 2015 Apr 29. EMBO J. 2015. PMID: 25925386 Free PMC article. Review. - The promises and challenges of human brain organoids as models of neuropsychiatric disease.
Quadrato G, Brown J, Arlotta P. Quadrato G, et al. Nat Med. 2016 Nov;22(11):1220-1228. doi: 10.1038/nm.4214. Epub 2016 Oct 26. Nat Med. 2016. PMID: 27783065 - Induced pluripotent stem cells: the new patient?
Bellin M, Marchetto MC, Gage FH, Mummery CL. Bellin M, et al. Nat Rev Mol Cell Biol. 2012 Nov;13(11):713-26. doi: 10.1038/nrm3448. Epub 2012 Oct 4. Nat Rev Mol Cell Biol. 2012. PMID: 23034453 Review. - Regulation of multiple signaling pathways promotes the consistent expansion of human pancreatic progenitors in defined conditions.
Jarc L, Bandral M, Zanfrini E, Lesche M, Kufrin V, Sendra R, Pezzolla D, Giannios I, Khattak S, Neumann K, Ludwig B, Gavalas A. Jarc L, et al. Elife. 2024 Jan 5;12:RP89962. doi: 10.7554/eLife.89962. Elife. 2024. PMID: 38180318 Free PMC article. - Transcriptional buffering and 3'UTR lengthening are shaped during human neurodevelopment by shifts in mRNA stability and microRNA load.
Mufteev M, Rodrigues DC, Yuki KE, Narula A, Wei W, Piekna A, Liu J, Pasceri P, Rissland OS, Wilson MD, Ellis J. Mufteev M, et al. bioRxiv [Preprint]. 2023 Mar 1:2023.03.01.530249. doi: 10.1101/2023.03.01.530249. bioRxiv. 2023. PMID: 36909614 Free PMC article. Preprint.
References
- Chahrour M., Zoghbi H.Y. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56:422–437. doi:10.1016/j.neuron.2007.10.001. - DOI - PubMed
- Hagberg B., Aicardi J., Dias K., Ramos O. A progressive syndrome of autism, dementia, ataxia, and loss of purposeful hand use in girls: Rett's syndrome: report of 35 cases. Ann. Neurol. 1983;14:471–479. doi:10.1002/ana.410140412. - DOI - PubMed
- Amir R.E., Van den Veyver I.B., Wan M., Tran C.Q., Francke U., Zoghbi H.Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi:10.1038/13810. - DOI - PubMed
- Nan X., Meehan R.R., Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–4892. doi:10.1093/nar/21.21.4886. - DOI - PMC - PubMed
- Nan X., Ng H.H., Johnson C.A., Laherty C.D., Turner B.M., Eisenman R.N., Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi:10.1038/30764. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials