Ubiquitination of E3 ligases: self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms - PubMed (original) (raw)
Review
. 2011 Sep;18(9):1393-402.
doi: 10.1038/cdd.2011.16. Epub 2011 Mar 4.
Affiliations
- PMID: 21372847
- PMCID: PMC3178436
- DOI: 10.1038/cdd.2011.16
Review
Ubiquitination of E3 ligases: self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms
P de Bie et al. Cell Death Differ. 2011 Sep.
Abstract
Ubiquitin modification of many cellular proteins targets them for proteasomal degradation, but in addition can also serve non-proteolytic functions. Over the last years, a significant progress has been made in our understanding of how modification of the substrates of the ubiquitin system is regulated. However, little is known on how the ubiquitin system that is comprised of ∼1500 components is regulated. Here, we discuss how the biggest subfamily within the system, that of the E3 ubiquitin ligases that endow the system with its high specificity towards the numerous substrates, is regulated and in particular via self-regulation mediated by ubiquitin modification. Ligases can be targeted for degradation in a self-catalyzed manner, or through modification mediated by an external ligase(s). In addition, non-proteolytic functions of self-ubiquitination, for example activation of the ligase, of E3s are discussed.
Figures
Figure 1
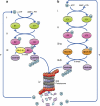
The ubiquitin-proteasome system. Conjugation of ubiquitin catalyzed by RING (a) or HECT (b) domain-containing ligases. (ai, bi) ATP-dependent activation of ubiquitin catalyzed by the ubiquitin-activating enzyme, E1. (aii, bii) Transfer of the activated ubiquitin to a ubiquitin-carrier protein (ubiquitin-conjugating enzyme), E2. In the case of a RING ligase, the ubiquitin-charged E2 binds to the E3 and transfers the activated ubiquitin moiety directly to the substrate that is also bound to the E3 (aiii). In the case of a HECT domain ligase, ubiquitin is transferred from the E2 to a Cys residue in the E3 (biii-a) and then to the substrate (biii-b). (iv and v) The conjugated substrate is degraded to short peptides by the 26S proteasome (iv) with release of free and reusable ubiquitin mediated by DUB(s) (v). Some of the ubiquitin is degraded in this process along with the substrate (iv)
Figure 2
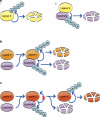
Self-ubiquitination and degradation of E3 ligases occur in three substrate-related modes: (a) Self-ubiquitination and degradation of the ligase (i) are independent from ubiquitination of the substrate (ii). (b) Self-ubiquitination occurs concomitantly with ubiquitination of the substrate. (c) Inhibition of self-ubiquitination by the substrate
Figure 3
Schematic representation of RING1B regulation by self-ubiquitination and ubiquitination by an exogenous ligase. RING1B is target to ubiquitination by E6-AP, and/or other E3 ligase(s), generating Lys48-based ubiquitin chains that target RING1B for proteasomal degradation. Self-ubiquitination of RING1B results in the formation of Lys6-, Lys27-, and Lys48-based mixed and multiply branched ubiquitin chains that activate it as a ligase for histone H2A. The balance between these two types of ubiquitination is regulated in several ways. (i) Since the activating and degrading forms of ubiquitination target the same lysine residues in RING1B, the two modifications are mutually exclusive. (ii) The PRC1 subunit BMI1 inhibits both types of ubiquitination. (iii) The DUB USP7 reverses both self- and E6-AP-mediated ubiquitination of RING1B, thereby returning it to its native state, potentially allowing the balance between the types of ubiquitination to be shifted. (iv) BMI1 stimulates ubiquitination of H2A by RING1B
Figure 4
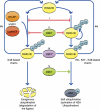
Schematic representation of the interplay between the ubiquitinating activities of APC/C and SCF complexes during cell cycle. (i) During G1 phase, APC/CCDH1 targets several F-box substrate receptors of SCF complexes, including SKP2 and TOME-1, and the SKP2 cofactor CKS1. In addition, CDC20 is also targeted to ubiquitination and degradation by APC/CCDH1. While CDH1 is subject to self-ubiquitination during G0 and G1 phases within the context of the APC/C complex (ii), the degradation of the free released phosphorylated form is mediated by an SCF complex during S phase (iii). (iv) During S and G2 phases, APC/CCDC20 is inhibited by the F-box protein EMI1 (iv-a); however, in the M phase, EMI1 is targeted for destruction by SCF_β_-TRCP alleviating the inhibition of APC/CCDC20 (iv-b). (v) CDC20 is targeted in a self-catalytic manner during the spindle-associated checkpoint
Figure 5
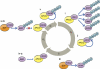
Mechanisms for proteasomal targeting of E3 ligases. (a) Degradation mediated by self-ubiquitination. (b) Hierarchical linear control of ligases. Ligase B is controlled by autoubiquitination (i) or another proteolytic mechanism, e.g., lysosomal degradation; (ii). It modifies in trans ligases C and D (iii), which in turn can modify ligases E and F, and ligases G, H, respectively (iv and v), etc. (c) Two (or more) ligases target one another in a closed circular manner
Similar articles
- p53 Ubiquitination and proteasomal degradation.
Love IM, Shi D, Grossman SR. Love IM, et al. Methods Mol Biol. 2013;962:63-73. doi: 10.1007/978-1-62703-236-0_5. Methods Mol Biol. 2013. PMID: 23150437 - Smad ubiquitylation regulatory factor 1/2 (Smurf1/2) promotes p53 degradation by stabilizing the E3 ligase MDM2.
Nie J, Xie P, Liu L, Xing G, Chang Z, Yin Y, Tian C, He F, Zhang L. Nie J, et al. J Biol Chem. 2010 Jul 23;285(30):22818-30. doi: 10.1074/jbc.M110.126920. Epub 2010 May 18. J Biol Chem. 2010. PMID: 20484049 Free PMC article. - HIV-1 Nef promotes ubiquitination and proteasomal degradation of p53 tumor suppressor protein by using E6AP.
Ali A, Farooqui SR, Rai J, Singh J, Kumar V, Mishra R, Banerjea AC. Ali A, et al. Biochem Biophys Res Commun. 2020 Sep 3;529(4):1038-1044. doi: 10.1016/j.bbrc.2020.05.188. Epub 2020 Jul 30. Biochem Biophys Res Commun. 2020. PMID: 32819562 - From Discovery to Bedside: Targeting the Ubiquitin System.
Wertz IE, Wang X. Wertz IE, et al. Cell Chem Biol. 2019 Feb 21;26(2):156-177. doi: 10.1016/j.chembiol.2018.10.022. Epub 2018 Dec 13. Cell Chem Biol. 2019. PMID: 30554913 Review. - Cullin-RING E3 Ubiquitin Ligases: Bridges to Destruction.
Nguyen HC, Wang W, Xiong Y. Nguyen HC, et al. Subcell Biochem. 2017;83:323-347. doi: 10.1007/978-3-319-46503-6_12. Subcell Biochem. 2017. PMID: 28271482 Free PMC article. Review.
Cited by
- Post-translational modification profiling - A novel tool for mapping the protein modification landscape in cancer.
Eisenberg-Lerner A, Ciechanover A, Merbl Y. Eisenberg-Lerner A, et al. Exp Biol Med (Maywood). 2016 Aug;241(14):1475-82. doi: 10.1177/1535370216651732. Epub 2016 May 26. Exp Biol Med (Maywood). 2016. PMID: 27229346 Free PMC article. Review. - Novel gene-intergenic fusion involving ubiquitin E3 ligase UBE3C causes distal hereditary motor neuropathy.
Cutrupi AN, Narayanan RK, Perez-Siles G, Grosz BR, Lai K, Boyling A, Ellis M, Lin RCY, Neumann B, Mao D, Uesugi M, Nicholson GA, Vucic S, Saporta MA, Kennerson ML. Cutrupi AN, et al. Brain. 2023 Mar 1;146(3):880-897. doi: 10.1093/brain/awac424. Brain. 2023. PMID: 36380488 Free PMC article. - The Role of NEDD4 E3 Ubiquitin-Protein Ligases in Parkinson's Disease.
Conway JA, Kinsman G, Kramer ER. Conway JA, et al. Genes (Basel). 2022 Mar 14;13(3):513. doi: 10.3390/genes13030513. Genes (Basel). 2022. PMID: 35328067 Free PMC article. Review. - Inhibition of USP28 overcomes Cisplatin-resistance of squamous tumors by suppression of the Fanconi anemia pathway.
Prieto-Garcia C, Hartmann O, Reissland M, Fischer T, Maier CR, Rosenfeldt M, Schülein-Völk C, Klann K, Kalb R, Dikic I, Münch C, Diefenbacher ME. Prieto-Garcia C, et al. Cell Death Differ. 2022 Mar;29(3):568-584. doi: 10.1038/s41418-021-00875-z. Epub 2021 Oct 5. Cell Death Differ. 2022. PMID: 34611298 Free PMC article. - E3 ubiquitin ligase-mediated regulation of bone formation and tumorigenesis.
Sévère N, Dieudonné FX, Marie PJ. Sévère N, et al. Cell Death Dis. 2013 Jan 17;4(1):e463. doi: 10.1038/cddis.2012.217. Cell Death Dis. 2013. PMID: 23328670 Free PMC article. Review.
References
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. - PubMed
- Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-CBL/SLI-1. Mol Cell. 1999;4:1029–1040. - PubMed
- Vodermaier HC. APC/C and SCF: controlling each other and the cell cycle. Curr Biol. 2004;14:R787–R796. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources